How Does Photoreceptor UVR8 Perceive a UV-B Signal?
- PMID: 25996910
- PMCID: PMC4560659
- DOI: 10.1111/php.12470
How Does Photoreceptor UVR8 Perceive a UV-B Signal?
Abstract
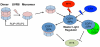
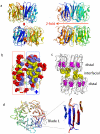
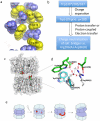
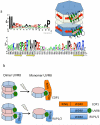
UVR8 is the only known plant photoreceptor that mediates light responses to UV-B (280-315 nm) of the solar spectrum. UVR8 perceives a UV-B signal via light-induced dimer dissociation, which triggers a wide range of cellular responses involved in photomorphogenesis and photoprotection. Two recent crystal structures of Arabidopsis thaliana UVR8 (AtUVR8) have revealed unusual clustering of UV-B-absorbing Trp pigments at the dimer interface and provided a structural framework for further mechanistic investigation. This review summarizes recent advances in spectroscopic, computational and crystallographic studies on UVR8 that are directed toward full understanding of UV-B perception at the molecular level.
© 2015 The American Society of Photobiology.
Figures


Similar articles
-
The UV-B photoreceptor UVR8: from structure to physiology.Plant Cell. 2014 Jan;26(1):21-37. doi: 10.1105/tpc.113.119446. Epub 2014 Jan 30. Plant Cell. 2014. PMID: 24481075 Free PMC article. Review.
-
Expression of Tomato UVR8 in Arabidopsis reveals conserved photoreceptor function.Plant Sci. 2021 Feb;303:110766. doi: 10.1016/j.plantsci.2020.110766. Epub 2020 Nov 21. Plant Sci. 2021. PMID: 33487351
-
Dimer/monomer status and in vivo function of salt-bridge mutants of the plant UV-B photoreceptor UVR8.Plant J. 2016 Oct;88(1):71-81. doi: 10.1111/tpj.13260. Epub 2016 Sep 9. Plant J. 2016. PMID: 27385642 Free PMC article.
-
The photoreceptor UVR8 mediates the perception of both UV-B and UV-A wavelengths up to 350 nm of sunlight with responsivity moderated by cryptochromes.Plant Cell Environ. 2020 Jun;43(6):1513-1527. doi: 10.1111/pce.13752. Epub 2020 Mar 24. Plant Cell Environ. 2020. PMID: 32167576
-
Recent advances in UV-B signalling: interaction of proteins with the UVR8 photoreceptor.J Exp Bot. 2025 Feb 7;76(3):873-881. doi: 10.1093/jxb/erae132. J Exp Bot. 2025. PMID: 38525857 Free PMC article. Review.
Cited by
-
UV-B light and its application potential to reduce disease and pest incidence in crops.Hortic Res. 2021 Sep 1;8(1):194. doi: 10.1038/s41438-021-00629-5. Hortic Res. 2021. PMID: 34465753 Free PMC article. Review.
-
Proline 411 biases the conformation of the intrinsically disordered plant UVR8 photoreceptor C27 domain altering the functional properties of the peptide.Sci Rep. 2019 Jan 28;9(1):818. doi: 10.1038/s41598-018-37005-8. Sci Rep. 2019. PMID: 30692548 Free PMC article.
-
Enhancement of germination and yield of cotton through optical seed priming: Lab. and diverse environment studies.PLoS One. 2023 Jul 20;18(7):e0288255. doi: 10.1371/journal.pone.0288255. eCollection 2023. PLoS One. 2023. PMID: 37471373 Free PMC article.
-
RUP2 facilitates UVR8 redimerization via two interfaces.Plant Commun. 2023 Jan 9;4(1):100428. doi: 10.1016/j.xplc.2022.100428. Epub 2022 Sep 5. Plant Commun. 2023. PMID: 36065466 Free PMC article.
-
UV-B Induces Chloroplast Movements in a Phototropin-Dependent Manner.Front Plant Sci. 2019 Oct 15;10:1279. doi: 10.3389/fpls.2019.01279. eCollection 2019. Front Plant Sci. 2019. PMID: 31681376 Free PMC article.
References
-
- Möglich A, Yang X, Ayers RA, Moffat K. Structure and Function of Plant Photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. - PubMed
-
- Rizzini L, Favory J-J, Cloix C, et al. Perception of UV-B by the Arabidopsis UVR8 Protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. - PubMed
-
- Wu D, Hu Q, Yan Z, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

