Context-Dependent Role of Mitochondrial Fusion-Fission in Clonal Expansion of mtDNA Mutations
- PMID: 25996936
- PMCID: PMC4440705
- DOI: 10.1371/journal.pcbi.1004183
Context-Dependent Role of Mitochondrial Fusion-Fission in Clonal Expansion of mtDNA Mutations
Abstract
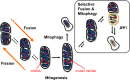
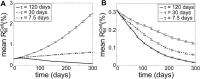
The accumulation of mutant mitochondrial DNA (mtDNA) molecules in aged cells has been associated with mitochondrial dysfunction, age-related diseases and the ageing process itself. This accumulation has been shown to often occur clonally, where mutant mtDNA grow in number and overpopulate the wild-type mtDNA. However, the cell possesses quality control (QC) mechanisms that maintain mitochondrial function, in which dysfunctional mitochondria are isolated and removed by selective fusion and mitochondrial autophagy (mitophagy), respectively. The aim of this study is to elucidate the circumstances related to mitochondrial QC that allow the expansion of mutant mtDNA molecules. For the purpose of the study, we have developed a mathematical model of mitochondrial QC process by extending our previous validated model of mitochondrial turnover and fusion-fission. A global sensitivity analysis of the model suggested that the selectivity of mitophagy and fusion is the most critical QC parameter for clearing de novo mutant mtDNA molecules. We further simulated several scenarios involving perturbations of key QC parameters to gain a better understanding of their dynamic and synergistic interactions. Our model simulations showed that a higher frequency of mitochondrial fusion-fission can provide a faster clearance of mutant mtDNA, but only when mutant-rich mitochondria that are transiently created are efficiently prevented from re-fusing with other mitochondria and selectively removed. Otherwise, faster fusion-fission quickens the accumulation of mutant mtDNA. Finally, we used the insights gained from model simulations and analysis to propose a possible circumstance involving deterioration of mitochondrial QC that permits mutant mtDNA to expand with age.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

