Identification of the Adapter Molecule MTSS1 as a Potential Oncogene-Specific Tumor Suppressor in Acute Myeloid Leukemia
- PMID: 25996952
- PMCID: PMC4440712
- DOI: 10.1371/journal.pone.0125783
Identification of the Adapter Molecule MTSS1 as a Potential Oncogene-Specific Tumor Suppressor in Acute Myeloid Leukemia
Abstract
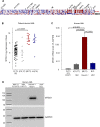
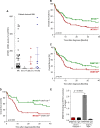
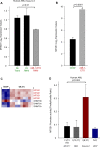
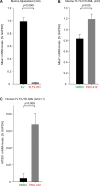
The adapter protein metastasis suppressor 1 (MTSS1) is implicated as a tumor suppressor or tumor promoter, depending on the type of solid cancer. Here, we identified Mtss1 expression to be increased in AML subsets with favorable outcome, while suppressed in high risk AML patients. High expression of MTSS1 predicted better clinical outcome of patients with normal-karyotype AML. Mechanistically, MTSS1 expression was negatively regulated by FLT3-ITD signaling but enhanced by the AML1-ETO fusion protein. DNMT3B, a negative regulator of MTSS1, showed strong binding to the MTSS1 promoter in PML-RARA positive but not AML1-ETO positive cells, suggesting that AML1-ETO leads to derepression of MTSS1. Pharmacological treatment of AML cell lines carrying the FLT3-ITD mutation with the specific FLT3 inhibitor PKC-412 caused upregulation of MTSS1. Moreover, treatment of acute promyelocytic cells (APL) with all-trans retinoic acid (ATRA) increased MTSS1 mRNA levels. Taken together, our findings suggest that MTSS1 might have a context-dependent function and could act as a tumor suppressor, which is pharmacologically targetable in AML patients.
Conflict of interest statement
Figures

Similar articles
-
AML1-ETO triggers epigenetic activation of early growth response gene l, inducing apoptosis in t(8;21) acute myeloid leukemia.FEBS J. 2014 Feb;281(4):1123-31. doi: 10.1111/febs.12673. Epub 2014 Jan 10. FEBS J. 2014. PMID: 24314118
-
FLT3-TKD mutation in childhood acute myeloid leukemia.Leukemia. 2003 May;17(5):883-6. doi: 10.1038/sj.leu.2402928. Leukemia. 2003. PMID: 12750701
-
Haematological & molecular profile of acute myelogenous leukaemia in India.Indian J Med Res. 2009 Mar;129(3):256-61. Indian J Med Res. 2009. PMID: 19491417
-
[THE CLINICAL SIGNIFICANCE OF GENETIC MUTATIONS IN ACUTE MYELOID LEUKEMIA].Lik Sprava. 2014 Dec;(12):10-8. Lik Sprava. 2014. PMID: 26638462 Review. Ukrainian.
-
The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias.Haematologica. 1997 May-Jun;82(3):364-70. Haematologica. 1997. PMID: 9234595 Review.
Cited by
-
Differential roles of STAT1 and STAT2 in the sensitivity of JAK2V617F- vs. BCR-ABL-positive cells to interferon alpha.J Hematol Oncol. 2019 Apr 2;12(1):36. doi: 10.1186/s13045-019-0722-9. J Hematol Oncol. 2019. PMID: 30940163 Free PMC article.
-
Emerging DNA Methylome Targets in FLT3-ITD-Positive Acute Myeloid Leukemia: Combination Therapy with Clinically Approved FLT3 Inhibitors.Curr Treat Options Oncol. 2024 Jun;25(6):719-751. doi: 10.1007/s11864-024-01202-7. Epub 2024 May 2. Curr Treat Options Oncol. 2024. PMID: 38696033 Free PMC article. Review.
-
MTSS1: beyond the integration of actin and membrane dynamics.Cell Mol Life Sci. 2024 Dec 3;81(1):472. doi: 10.1007/s00018-024-05511-w. Cell Mol Life Sci. 2024. PMID: 39625546 Free PMC article. Review.
-
Metastasis suppressor genes in clinical practice: are they druggable?Cancer Metastasis Rev. 2023 Dec;42(4):1169-1188. doi: 10.1007/s10555-023-10135-w. Epub 2023 Sep 25. Cancer Metastasis Rev. 2023. PMID: 37749308 Free PMC article. Review.
-
Mtss1 is a critical epigenetically regulated tumor suppressor in CML.Leukemia. 2016 Apr;30(4):823-32. doi: 10.1038/leu.2015.329. Epub 2015 Dec 1. Leukemia. 2016. PMID: 26621336
References
-
- Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;8:1586–95. - PubMed
-
- Brunet S, Labopin M, Esteve J, Cornelissen J, Socié G, Iori AP, et al. Impact of FLT3 internal tandem duplication on the outcome of related and unrelated hematopoietic transplantation for adult acute myeloid leukemia in first remission: a retrospective analysis. J Clin Oncol. 2012; 1;30(7):735–41. 10.1200/JCO.2011.36.9868 - DOI - PubMed
-
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010; 116(3):354–65. 10.1182/blood-2009-11-254441 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

