DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation
- PMID: 25999370
- PMCID: PMC4937790
- DOI: 10.1126/science.aab1433
DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation
Abstract
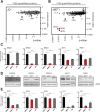
The development of effective pharmacological inhibitors of multidomain scaffold proteins, notably transcription factors, is a particularly challenging problem. In part, this is because many small-molecule antagonists disrupt the activity of only one domain in the target protein. We devised a chemical strategy that promotes ligand-dependent target protein degradation using as an example the transcriptional coactivator BRD4, a protein critical for cancer cell growth and survival. We appended a competitive antagonist of BET bromodomains to a phthalimide moiety to hijack the cereblon E3 ubiquitin ligase complex. The resultant compound, dBET1, induced highly selective cereblon-dependent BET protein degradation in vitro and in vivo and delayed leukemia progression in mice. A second series of probes resulted in selective degradation of the cytosolic protein FKBP12. This chemical strategy for controlling target protein stability may have implications for therapeutically targeting previously intractable proteins.
Copyright © 2015, American Association for the Advancement of Science.
Figures



Comment in
-
Anticancer drugs: Selectively targeting proteins for degradation.Nat Rev Drug Discov. 2015 Jul;14(7):459. doi: 10.1038/nrd4670. Epub 2015 Jun 19. Nat Rev Drug Discov. 2015. PMID: 26091269 No abstract available.
-
Protein-slaying drugs could be the next blockbuster therapies.Nature. 2019 Mar;567(7748):298-300. doi: 10.1038/d41586-019-00879-3. Nature. 2019. PMID: 30894734 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

