Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin
- PMID: 25999502
- PMCID: PMC4471149
- DOI: 10.1126/science.aaa6806
Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin
Abstract
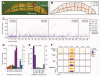
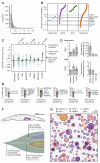
How somatic mutations accumulate in normal cells is central to understanding cancer development but is poorly understood. We performed ultradeep sequencing of 74 cancer genes in small (0.8 to 4.7 square millimeters) biopsies of normal skin. Across 234 biopsies of sun-exposed eyelid epidermis from four individuals, the burden of somatic mutations averaged two to six mutations per megabase per cell, similar to that seen in many cancers, and exhibited characteristic signatures of exposure to ultraviolet light. Remarkably, multiple cancer genes are under strong positive selection even in physiologically normal skin, including most of the key drivers of cutaneous squamous cell carcinomas. Positively selected mutations were found in 18 to 32% of normal skin cells at a density of ~140 driver mutations per square centimeter. We observed variability in the driver landscape among individuals and variability in the sizes of clonal expansions across genes. Thus, aged sun-exposed skin is a patchwork of thousands of evolving clones with over a quarter of cells carrying cancer-causing mutations while maintaining the physiological functions of epidermis.
Copyright © 2015, American Association for the Advancement of Science.
Figures


Comment in
-
Cancer. Preprocancer.Science. 2015 May 22;348(6237):867-8. doi: 10.1126/science.aac4435. Science. 2015. PMID: 25999495 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

