Ecological prevalence, genetic diversity, and epidemiological aspects of Salmonella isolated from tomato agricultural regions of the Virginia Eastern Shore
- PMID: 25999938
- PMCID: PMC4423467
- DOI: 10.3389/fmicb.2015.00415
Ecological prevalence, genetic diversity, and epidemiological aspects of Salmonella isolated from tomato agricultural regions of the Virginia Eastern Shore
Abstract
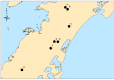
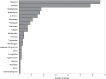
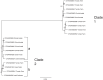
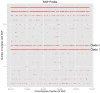
Virginia is the third largest producer of fresh-market tomatoes in the United States. Tomatoes grown along the eastern shore of Virginia are implicated almost yearly in Salmonella illnesses. Traceback implicates contamination occurring in the pre-harvest environment. To get a better understanding of the ecological niches of Salmonella in the tomato agricultural environment, a 2-year study was undertaken at a regional agricultural research farm in Virginia. Environmental samples, including tomato (fruit, blossoms, and leaves), irrigation water, surface water and sediment, were collected over the growing season. These samples were analyzed for the presence of Salmonella using modified FDA-BAM methods. Molecular assays were used to screen the samples. Over 1500 samples were tested. Seventy-five samples tested positive for Salmonella yielding over 230 isolates. The most commonly isolated serovars were S. Newport and S. Javiana with pulsed-field gel electrophoresis yielding 39 different patterns. Genetic diversity was further underscored among many other serotypes, which showed multiple PFGE subtypes. Whole genome sequencing (WGS) of several S. Newport isolates collected in 2010 compared to clinical isolates associated with tomato consumption showed very few single nucleotide differences between environmental isolates and clinical isolates suggesting a source link to Salmonella contaminated tomatoes. Nearly all isolates collected during two growing seasons of surveillance were obtained from surface water and sediment sources pointing to these sites as long-term reservoirs for persistent and endemic contamination of this environment.
Keywords: Salmonella Newport; environmental reservoirs; epidemiological impact; prevalence and diversity; tomatoes.
Figures


References
-
- Andrews W. H., Jacobson A., Hammack T. S. (2011). “Salmonella,” in Bacteriological Analytical Manual (U.S. Food and Drug Administration). Available online at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

