Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress
- PMID: 25999972
- PMCID: PMC4422006
- DOI: 10.3389/fpls.2015.00308
Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress
Abstract
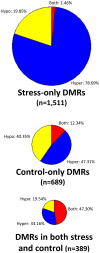
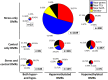
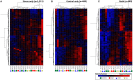
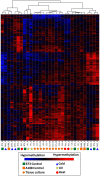
DNA methylation is a chromatin modification that is sometimes associated with epigenetic regulation of gene expression. As DNA methylation can be reversible at some loci, it is possible that methylation patterns may change within an organism that is subjected to environmental stress. In order to assess the effects of abiotic stress on DNA methylation patterns in maize (Zea mays), seeding plants were subjected to heat, cold, and UV stress treatments. Tissue was later collected from individual adult plants that had been subjected to stress or control treatments and used to perform DNA methylation profiling to determine whether there were consistent changes in DNA methylation triggered by specific stress treatments. DNA methylation profiling was performed by immunoprecipitation of methylated DNA followed by microarray hybridization to allow for quantitative estimates of DNA methylation abundance throughout the low-copy portion of the maize genome. By comparing the DNA methylation profiles of each individual plant to the average of the control plants it was possible to identify regions of the genome with variable DNA methylation. However, we did not find evidence of consistent DNA methylation changes resulting from the stress treatments used in this study. Instead, the data suggest that there is a low-rate of stochastic variation that is present in both control and stressed plants.
Keywords: DNA methylation; abiotic stress; epigenetics; maize; tissue culture.
Figures

Similar articles
-
Heritable Epigenomic Changes to the Maize Methylome Resulting from Tissue Culture.Genetics. 2018 Aug;209(4):983-995. doi: 10.1534/genetics.118.300987. Epub 2018 May 30. Genetics. 2018. PMID: 29848487 Free PMC article.
-
The dynamics of DNA methylation in maize roots under Pb stress.Int J Mol Sci. 2014 Dec 17;15(12):23537-54. doi: 10.3390/ijms151223537. Int J Mol Sci. 2014. PMID: 25526567 Free PMC article.
-
Consistent and heritable alterations of DNA methylation are induced by tissue culture in maize.Genetics. 2014 Sep;198(1):209-18. doi: 10.1534/genetics.114.165480. Epub 2014 Jul 14. Genetics. 2014. PMID: 25023398 Free PMC article.
-
Epigenetic Control of Gene Expression in Maize.Int Rev Cell Mol Biol. 2017;328:25-48. doi: 10.1016/bs.ircmb.2016.08.002. Epub 2016 Sep 28. Int Rev Cell Mol Biol. 2017. PMID: 28069135 Review.
-
Retrospective and perspective of plant epigenetics in China.J Genet Genomics. 2018 Nov 20;45(11):621-638. doi: 10.1016/j.jgg.2018.09.004. Epub 2018 Nov 6. J Genet Genomics. 2018. PMID: 30455036 Review.
Cited by
-
Whole genome bisulfite sequencing methylome analysis of mulberry (Morus alba) reveals epigenome modifications in response to drought stress.Sci Rep. 2020 May 15;10(1):8013. doi: 10.1038/s41598-020-64975-5. Sci Rep. 2020. PMID: 32415195 Free PMC article.
-
DNA methylation analysis in plants: review of computational tools and future perspectives.Brief Bioinform. 2020 May 21;21(3):906-918. doi: 10.1093/bib/bbz039. Brief Bioinform. 2020. PMID: 31220217 Free PMC article. Review.
-
Genome-wide DNA methylation and their transgenerational pattern differ in Arabidopsis thaliana populations originated along the elevation of West Himalaya.BMC Plant Biol. 2024 Oct 9;24(1):936. doi: 10.1186/s12870-024-05641-0. BMC Plant Biol. 2024. PMID: 39385079 Free PMC article.
-
Stable unmethylated DNA demarcates expressed genes and their cis-regulatory space in plant genomes.Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23991-24000. doi: 10.1073/pnas.2010250117. Epub 2020 Sep 2. Proc Natl Acad Sci U S A. 2020. PMID: 32879011 Free PMC article.
-
Global Methylomic and Transcriptomic Analyses Reveal the Broad Participation of DNA Methylation in Daily Gene Expression Regulation of Populus trichocarpa.Front Plant Sci. 2019 Feb 28;10:243. doi: 10.3389/fpls.2019.00243. eCollection 2019. Front Plant Sci. 2019. PMID: 30873202 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

