Using Genome-scale Models to Predict Biological Capabilities
- PMID: 26000478
- PMCID: PMC4451052
- DOI: 10.1016/j.cell.2015.05.019
Using Genome-scale Models to Predict Biological Capabilities
Abstract
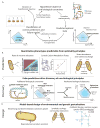
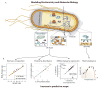
Constraint-based reconstruction and analysis (COBRA) methods at the genome scale have been under development since the first whole-genome sequences appeared in the mid-1990s. A few years ago, this approach began to demonstrate the ability to predict a range of cellular functions, including cellular growth capabilities on various substrates and the effect of gene knockouts at the genome scale. Thus, much interest has developed in understanding and applying these methods to areas such as metabolic engineering, antibiotic design, and organismal and enzyme evolution. This Primer will get you started.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

