Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles
- PMID: 26000485
- PMCID: PMC4448932
- DOI: 10.1016/j.cell.2015.04.030
Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles
Abstract
Mammalian mitotic chromosome morphogenesis was analyzed by 4D live-cell and snapshot deconvolution fluorescence imaging. Prophase chromosomes, whose organization was previously unknown, are revealed to comprise co-oriented sister linear loop arrays displayed along a single, peripheral, regularly kinked topoisomerase II/cohesin/condensin II axis. Thereafter, rather than smooth, progressive compaction as generally envisioned, progression to metaphase is a discontinuous process involving chromosome expansion as well as compaction. At late prophase, dependent on topoisomerase II and with concomitant cohesin release, chromosomes expand, axes split and straighten, and chromatin loops transit to a radial disposition around now-central axes. Finally, chromosomes globally compact, giving the metaphase state. These patterns are consistent with the hypothesis that the molecular events of chromosome morphogenesis are governed by accumulation and release of chromosome stress, created by chromatin compaction and expansion. Chromosome state could evolve analogously throughout the cell cycle.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


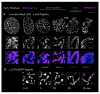


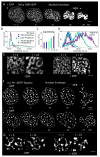

References
-
- Almagro S, Riveline D, Hirano T, Houchmandzadeh B, Dimitrov S. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J Biol Chem. 2004;279:5118–26. - PubMed
-
- Bajer A. Changes in length and volume of mitotic chromosomes in living cells. Hereditas. 1959;45:579–596.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

