The cerebellum ages slowly according to the epigenetic clock
- PMID: 26000617
- PMCID: PMC4468311
- DOI: 10.18632/aging.100742
The cerebellum ages slowly according to the epigenetic clock
Abstract
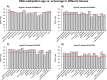
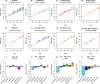
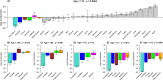
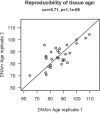
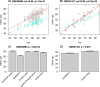
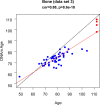
Studies that elucidate why some human tissues age faster than others may shed light on how we age, and ultimately suggest what interventions may be possible. Here we utilize a recent biomarker of aging (referred to as epigenetic clock) to assess the epigenetic ages of up to 30 anatomic sites from supercentenarians (subjects who reached an age of 110 or older) and younger subjects. Using three novel and three published human DNA methylation data sets, we demonstrate that the cerebellum ages more slowly than other parts of the human body. We used both transcriptional data and genetic data to elucidate molecular mechanisms which may explain this finding. The two largest superfamilies of helicases (SF1 and SF2) are significantly over-represented (p=9.2x10-9) among gene transcripts that are over-expressed in the cerebellum compared to other brain regions from the same subject. Furthermore, SNPs that are associated with epigenetic age acceleration in the cerebellum tend to be located near genes from helicase superfamilies SF1 and SF2 (enrichment p=5.8x10-3). Our genetic and transcriptional studies of epigenetic age acceleration support the hypothesis that the slow aging rate of the cerebellum is due to processes that involve RNA helicases.
Keywords: biomarker of aging; brain; centenarian; epigenetics; tissue aging.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Baker G, Sprott R. Biomarkers of aging. Exp Gerontol. 1988;23:223–239. - PubMed
-
- Sanders J, Boudreau R. Dordrecht Heidelberg. New York: Springer; 2012. Understanding the Aging Process Using Epidemiologic Approaches.
-
- Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
