The centrosomal linker and microtubules provide dual levels of spatial coordination of centrosomes
- PMID: 26001056
- PMCID: PMC4441491
- DOI: 10.1371/journal.pgen.1005243
The centrosomal linker and microtubules provide dual levels of spatial coordination of centrosomes
Abstract
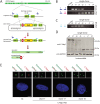
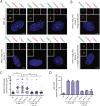
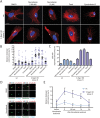
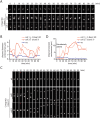
The centrosome is the principal microtubule organizing center in most animal cells. It consists of a pair of centrioles surrounded by pericentriolar material. The centrosome, like DNA, duplicates exactly once per cell cycle. During interphase duplicated centrosomes remain closely linked by a proteinaceous linker. This centrosomal linker is composed of rootletin filaments that are anchored to the centrioles via the protein C-Nap1. At the onset of mitosis the linker is dissolved by Nek2A kinase to support the formation of the bipolar mitotic spindle. The importance of the centrosomal linker for cell function during interphase awaits characterization. Here we assessed the phenotype of human RPE1 C-Nap1 knockout (KO) cells. The absence of the linker led to a modest increase in the average centrosome separation from 1 to 2.5 μm. This small impact on the degree of separation is indicative of a second level of spatial organization of centrosomes. Microtubule depolymerisation or stabilization in C-Nap1 KO cells dramatically increased the inter-centrosomal separation (> 8 μm). Thus, microtubules position centrosomes relatively close to one another in the absence of linker function. C-Nap1 KO cells had a Golgi organization defect with a two-fold expansion of the area occupied by the Golgi. When the centrosomes of C-Nap1 KO cells showed considerable separation, two spatially distinct Golgi stacks could be observed. Furthermore, migration of C-Nap1 KO cells was slower than their wild type RPE1 counterparts. These data show that the spatial organization of centrosomes is modulated by a combination of centrosomal cohesion and microtubule forces. Furthermore a modest increase in centrosome separation has major impact on Golgi organization and cell migration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Centrosome linker diversity and its function in centrosome clustering and mitotic spindle formation.EMBO J. 2023 Sep 4;42(17):e109738. doi: 10.15252/embj.2021109738. Epub 2023 Jul 4. EMBO J. 2023. PMID: 37401899 Free PMC article.
-
LRRC45 is a centrosome linker component required for centrosome cohesion.Cell Rep. 2013 Sep 26;4(6):1100-7. doi: 10.1016/j.celrep.2013.08.005. Epub 2013 Sep 12. Cell Rep. 2013. PMID: 24035387
-
The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles.J Cell Sci. 2002 Aug 15;115(Pt 16):3275-84. doi: 10.1242/jcs.115.16.3275. J Cell Sci. 2002. PMID: 12140259
-
Separate to operate: control of centrosome positioning and separation.Philos Trans R Soc Lond B Biol Sci. 2014 Sep 5;369(1650):20130461. doi: 10.1098/rstb.2013.0461. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25047615 Free PMC article. Review.
-
Emerging roles of centrosome cohesion.Open Biol. 2022 Oct;12(10):220229. doi: 10.1098/rsob.220229. Epub 2022 Oct 26. Open Biol. 2022. PMID: 36285440 Free PMC article. Review.
Cited by
-
Stable centrosomal roots disentangle to allow interphase centriole independence.PLoS Biol. 2018 Apr 12;16(4):e2003998. doi: 10.1371/journal.pbio.2003998. eCollection 2018 Apr. PLoS Biol. 2018. Update in: PLoS Biol. 2022 Oct 25;20(10):e3001854. doi: 10.1371/journal.pbio.3001854. PMID: 29649211 Free PMC article. Updated.
-
Centrosome linker diversity and its function in centrosome clustering and mitotic spindle formation.EMBO J. 2023 Sep 4;42(17):e109738. doi: 10.15252/embj.2021109738. Epub 2023 Jul 4. EMBO J. 2023. PMID: 37401899 Free PMC article.
-
Bystanders or active players: the role of extra centrosomes as signaling hubs.Cancer Metastasis Rev. 2024 Nov 20;44(1):1. doi: 10.1007/s10555-024-10224-4. Cancer Metastasis Rev. 2024. PMID: 39570514 Free PMC article. Review.
-
STED nanoscopy of the centrosome linker reveals a CEP68-organized, periodic rootletin network anchored to a C-Nap1 ring at centrioles.Proc Natl Acad Sci U S A. 2018 Mar 6;115(10):E2246-E2253. doi: 10.1073/pnas.1716840115. Epub 2018 Feb 20. Proc Natl Acad Sci U S A. 2018. PMID: 29463719 Free PMC article.
-
Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia.Dev Cell. 2016 Nov 21;39(4):424-437. doi: 10.1016/j.devcel.2016.10.006. Epub 2016 Nov 3. Dev Cell. 2016. PMID: 27818179 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

