ATRX represses alternative lengthening of telomeres
- PMID: 26001292
- PMCID: PMC4599288
- DOI: 10.18632/oncotarget.3846
ATRX represses alternative lengthening of telomeres
Abstract
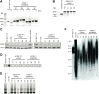
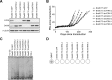
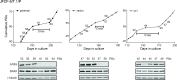
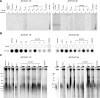
The unlimited proliferation of cancer cells requires a mechanism to prevent telomere shortening. Alternative Lengthening of Telomeres (ALT) is an homologous recombination-mediated mechanism of telomere elongation used in tumors, including osteosarcomas, soft tissue sarcoma subtypes, and glial brain tumors. Mutations in the ATRX/DAXX chromatin remodeling complex have been reported in tumors and cell lines that use the ALT mechanism, suggesting that ATRX may be an ALT repressor. We show here that knockout or knockdown of ATRX in mortal cells or immortal telomerase-positive cells is insufficient to activate ALT. Notably, however, in SV40-transformed mortal fibroblasts ATRX loss results in either a significant increase in the proportion of cell lines activating ALT (instead of telomerase) or in a significant decrease in the time prior to ALT activation. These data indicate that loss of ATRX function cooperates with one or more as-yet unidentified genetic or epigenetic alterations to activate ALT. Moreover, transient ATRX expression in ALT-positive/ATRX-negative cells represses ALT activity. These data provide the first direct, functional evidence that ATRX represses ALT.
Keywords: ALT; ATRX; immortalization; telomere.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures


References
-
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. - PubMed
-
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. - PubMed
-
- Perrem K, Bryan TM, Englezou A, Hackl T, Moy EL, Reddel RR. Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene. 1999;18:3383–3390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

