High throughput profiling of the cotton bollworm Helicoverpa armigera immunotranscriptome during the fungal and bacterial infections
- PMID: 26001831
- PMCID: PMC4490664
- DOI: 10.1186/s12864-015-1509-1
High throughput profiling of the cotton bollworm Helicoverpa armigera immunotranscriptome during the fungal and bacterial infections
Abstract
Background: Innate immunity is essential in defending against invading pathogens in invertebrates. The cotton bollworm, Helicoverpa armigera (Hübner) is one of the most destructive lepidopteran pests, which causes enormous economic losses in agricultural production worldwide. The components of the immune system are largely unknown in this insect. The application of entomopathogens is considered as an alternative to the chemical insecticides for its control. However, few studies have focused on the molecular mechanisms of host-pathogen interactions between pest insects and their pathogens. Here, we investigated the immunotranscriptome of H. armigera larvae and examined gene expression changes after pathogen infections. This study provided insights into the potential immunity-related genes and pathways in H. armigera larvae.
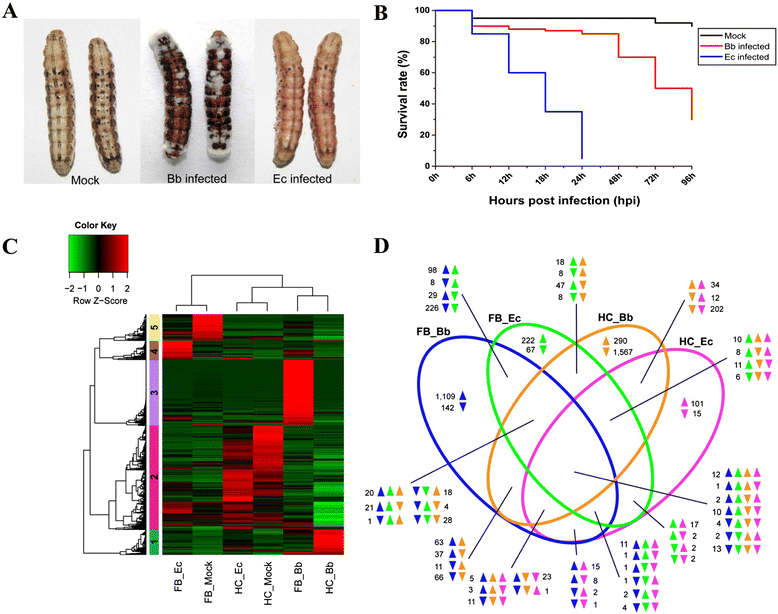
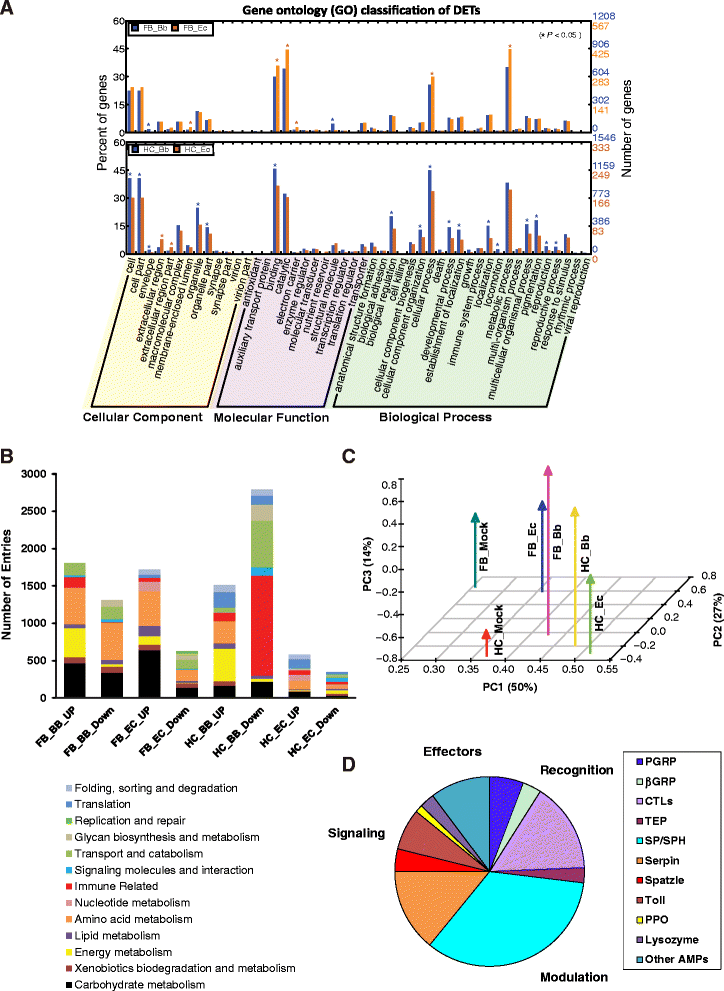
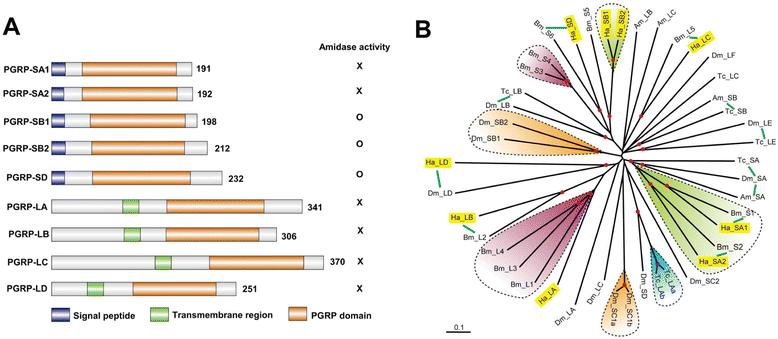
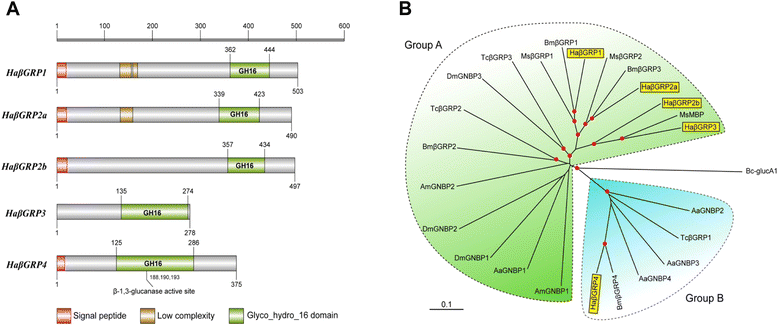
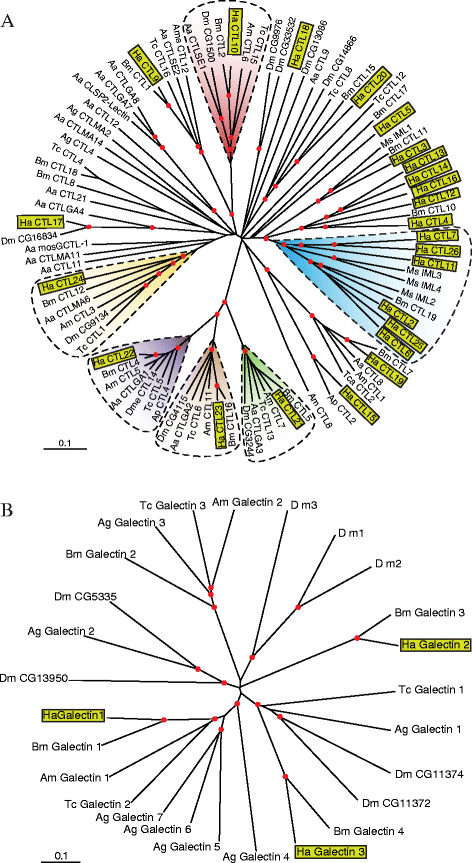
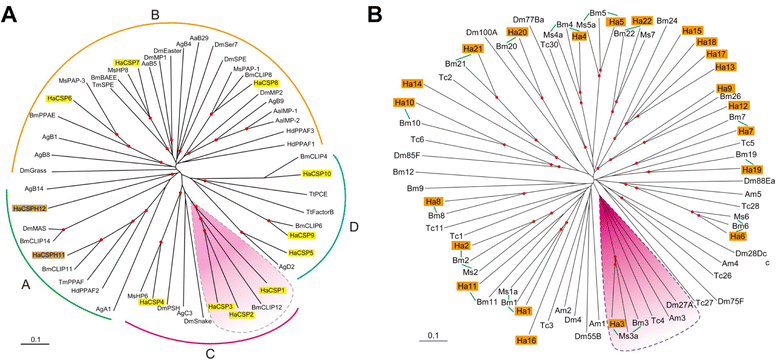
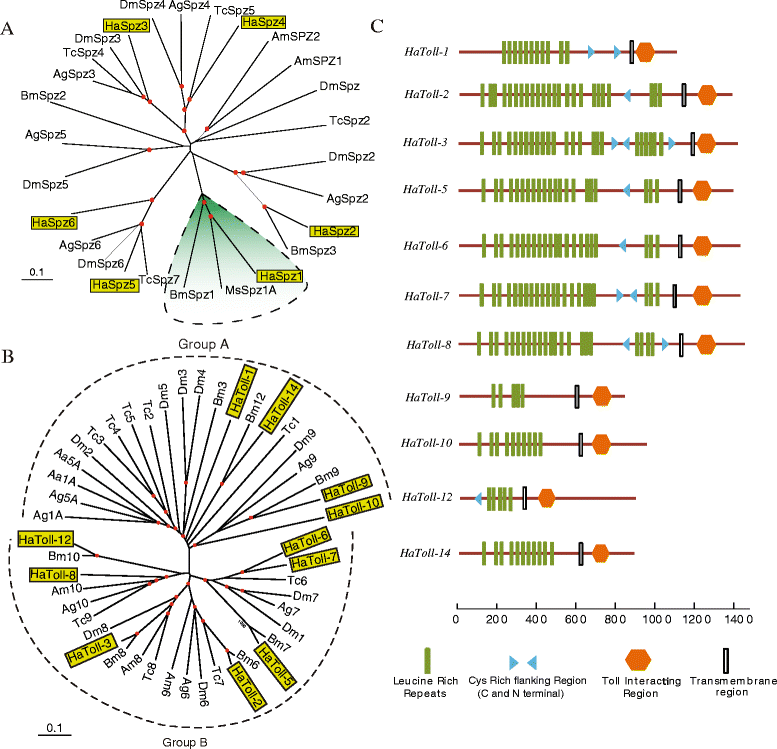
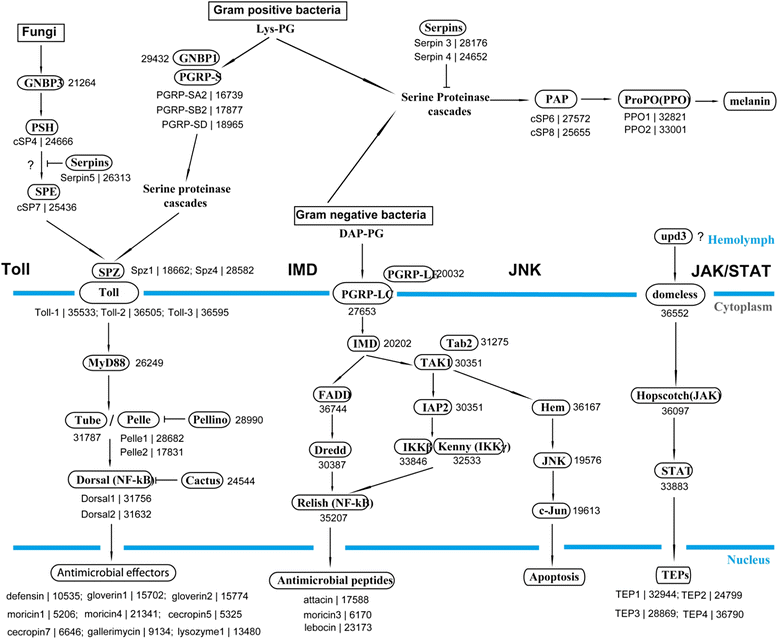
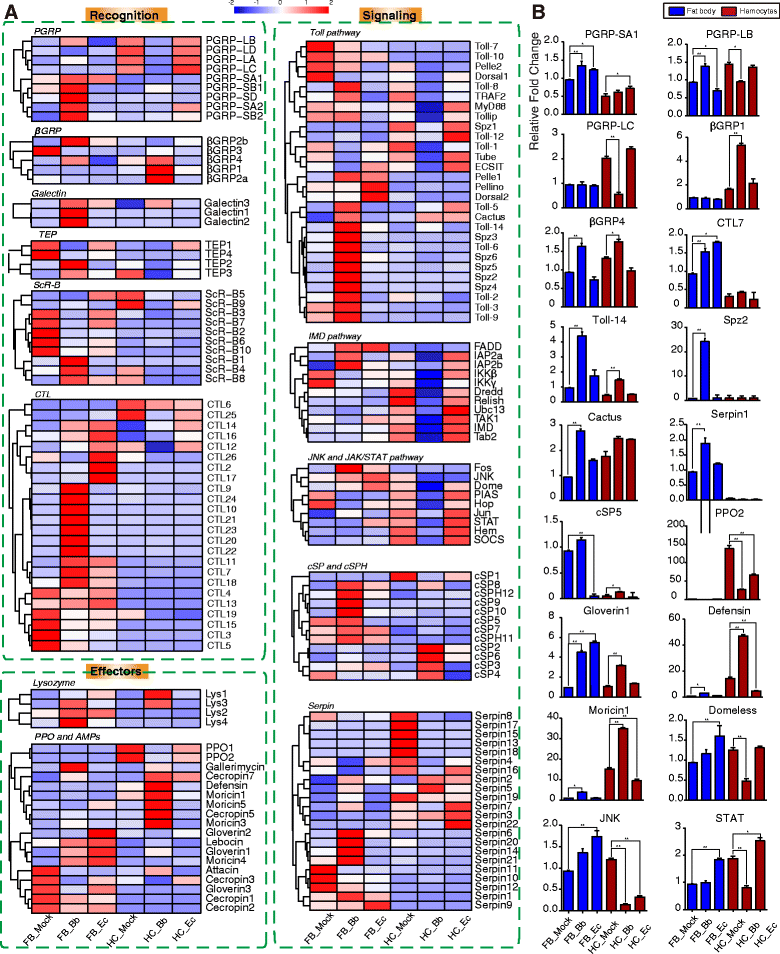
Results: Here, we adopted a high throughput RNA-seq approach to determine the immunotranscriptome of H. armigera larvae injected with buffer, fungal pathogen Beauveria bassiana, or Gram-negative bacterium Enterobacter cloacae. Based on sequence similarity to those homologs known to participate in immune responses in other insects, we identified immunity-related genes encoding pattern recognition receptors, signal modulators, immune effectors, and nearly all members of the Toll, IMD and JAK/STAT pathways. The RNA-seq data indicated that some immunity-related genes were activated in fungus- and bacterium-challenged fat body while others were suppressed in B. bassiana challenged hemocytes, including the putative IMD and JAK-STAT pathway members. Bacterial infection elevated the expression of recognition and modulator genes in the fat body and signal pathway genes in hemocytes. Although fat body and hemocytes both are important organs involved in the immune response, our transcriptome analysis revealed that more immunity-related genes were induced in the fat body than that hemocytes. Furthermore, quantitative real-time PCR analysis confirmed that, consistent with the RNA-seq data, the transcript abundances of putative PGRP-SA1, Serpin1, Toll-14, and Spz2 genes were elevated in fat body upon B. bassiana infection, while the mRNA levels of defensin, moricin1, and gloverin1 were up-regulated in hemocytes.
Conclusions: In this study, a global survey of the host defense against fungal and bacterial infection was performed on the non-model lepidopteran pest species. The comprehensive sequence resource and expression profiles of the immunity-related genes in H. armigera are acquired. This study provided valuable information for future functional investigations as well as development of specific and effective agents to control this pest.
Figures
Similar articles
-
Molecular characterization of a peptidoglycan recognition protein from the cotton bollworm, Helicoverpa armigera and its role in the prophenoloxidase activation pathway.Mol Immunol. 2015 May;65(1):123-32. doi: 10.1016/j.molimm.2015.01.016. Epub 2015 Feb 3. Mol Immunol. 2015. PMID: 25659083
-
20-hydroxyecdysone transcriptionally regulates humoral immunity in the fat body of Helicoverpa armigera.Insect Mol Biol. 2014 Dec;23(6):842-56. doi: 10.1111/imb.12131. Epub 2014 Sep 15. Insect Mol Biol. 2014. PMID: 25224836
-
Dynamics of the Interaction between Cotton Bollworm Helicoverpa armigera and Nucleopolyhedrovirus as Revealed by Integrated Transcriptomic and Proteomic Analyses.Mol Cell Proteomics. 2017 Jun;16(6):1009-1028. doi: 10.1074/mcp.M116.062547. Epub 2017 Apr 12. Mol Cell Proteomics. 2017. PMID: 28404795 Free PMC article.
-
Pattern recognition receptors from lepidopteran insects and their biological functions.Dev Comp Immunol. 2020 Jul;108:103688. doi: 10.1016/j.dci.2020.103688. Epub 2020 Mar 25. Dev Comp Immunol. 2020. PMID: 32222357 Review.
-
Baculovirus induced transcripts in hemocytes from the larvae of Heliothis virescens.Viruses. 2011 Nov;3(11):2047-64. doi: 10.3390/v3112047. Epub 2011 Oct 28. Viruses. 2011. PMID: 22163334 Free PMC article. Review.
Cited by
-
Dual roles and evolutionary implications of P26/poxin in antagonizing intracellular cGAS-STING and extracellular melanization immunity.Nat Commun. 2022 Nov 14;13(1):6934. doi: 10.1038/s41467-022-34761-0. Nat Commun. 2022. PMID: 36376305 Free PMC article.
-
A Genome-Wide Analysis of Serine Protease Inhibitors in Cydia pomonella Provides Insights into Their Evolution and Expression Pattern.Int J Mol Sci. 2023 Nov 15;24(22):16349. doi: 10.3390/ijms242216349. Int J Mol Sci. 2023. PMID: 38003538 Free PMC article.
-
Functional characterization of two clip domain serine proteases in innate immune responses of Aedes aegypti.Parasit Vectors. 2021 Nov 24;14(1):584. doi: 10.1186/s13071-021-05091-9. Parasit Vectors. 2021. PMID: 34819136 Free PMC article.
-
An entomopathogenic fungus exploits its host humoral antibacterial immunity to minimize bacterial competition in the hemolymph.Microbiome. 2023 May 20;11(1):116. doi: 10.1186/s40168-023-01538-6. Microbiome. 2023. PMID: 37210573 Free PMC article.
-
Comparative Proteomic Insights into the Immune Response of Conogethes punctiferalis Challenged with Beauveria bassiana.Insects. 2025 Jun 26;16(7):667. doi: 10.3390/insects16070667. Insects. 2025. PMID: 40725299 Free PMC article.
References
-
- Wang CS, Feng MG. Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol Control. 2014;68:129–35. doi: 10.1016/j.biocontrol.2013.06.017. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

