Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence
- PMID: 26002083
- PMCID: PMC4473727
- DOI: 10.7554/eLife.06792
Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence
Abstract
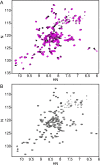
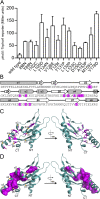
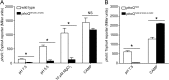
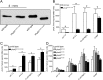
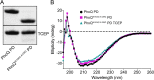
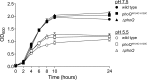
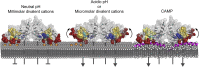
Salmonella PhoQ is a histidine kinase with a periplasmic sensor domain (PD) that promotes virulence by detecting the macrophage phagosome. PhoQ activity is repressed by divalent cations and induced in environments of acidic pH, limited divalent cations, and cationic antimicrobial peptides (CAMP). Previously, it was unclear which signals are sensed by salmonellae to promote PhoQ-mediated virulence. We defined conformational changes produced in the PhoQ PD on exposure to acidic pH that indicate structural flexibility is induced in α-helices 4 and 5, suggesting this region contributes to pH sensing. Therefore, we engineered a disulfide bond between W104C and A128C in the PhoQ PD that restrains conformational flexibility in α-helices 4 and 5. PhoQ(W104C-A128C) is responsive to CAMP, but is inhibited for activation by acidic pH and divalent cation limitation. phoQ(W104C-A128C) Salmonella enterica Typhimurium is virulent in mice, indicating that acidic pH and divalent cation sensing by PhoQ are dispensable for virulence.
Keywords: Salmonella; bacterial pathogenesis; biochemistry; infectious disease; innate immunity; intracellular virulence; microbiology; sensor kinase; signal transduction.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D, Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

