Genome-Wide Mapping of Structural Variations Reveals a Copy Number Variant That Determines Reproductive Morphology in Cucumber
- PMID: 26002866
- PMCID: PMC4498199
- DOI: 10.1105/tpc.114.135848
Genome-Wide Mapping of Structural Variations Reveals a Copy Number Variant That Determines Reproductive Morphology in Cucumber
Abstract
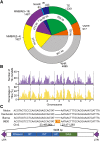
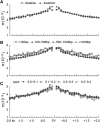
Structural variations (SVs) represent a major source of genetic diversity. However, the functional impact and formation mechanisms of SVs in plant genomes remain largely unexplored. Here, we report a nucleotide-resolution SV map of cucumber (Cucumis sativas) that comprises 26,788 SVs based on deep resequencing of 115 diverse accessions. The largest proportion of cucumber SVs was formed through nonhomologous end-joining rearrangements, and the occurrence of SVs is closely associated with regions of high nucleotide diversity. These SVs affect the coding regions of 1676 genes, some of which are associated with cucumber domestication. Based on the map, we discovered a copy number variation (CNV) involving four genes that defines the Female (F) locus and gives rise to gynoecious cucumber plants, which bear only female flowers and set fruit at almost every node. The CNV arose from a recent 30.2-kb duplication at a meiotically unstable region, likely via microhomology-mediated break-induced replication. The SV set provides a snapshot of structural variations in plants and will serve as an important resource for exploring genes underlying key traits and for facilitating practical breeding in cucumber.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures

Similar articles
-
Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber.Nat Commun. 2022 Feb 3;13(1):682. doi: 10.1038/s41467-022-28362-0. Nat Commun. 2022. PMID: 35115520 Free PMC article.
-
Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly.Plant J. 2012 Sep;71(6):895-906. doi: 10.1111/j.1365-313X.2012.05017.x. Epub 2012 Jul 9. Plant J. 2012. PMID: 22487099
-
A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity.Nat Genet. 2013 Dec;45(12):1510-5. doi: 10.1038/ng.2801. Epub 2013 Oct 20. Nat Genet. 2013. PMID: 24141363
-
Insights into structural variations and genome rearrangements in prokaryotic genomes.Bioinformatics. 2015 Jan 1;31(1):1-9. doi: 10.1093/bioinformatics/btu600. Epub 2014 Sep 4. Bioinformatics. 2015. PMID: 25189783 Review.
-
Molecular basis of cucumber fruit domestication.Curr Opin Plant Biol. 2019 Feb;47:38-46. doi: 10.1016/j.pbi.2018.08.006. Epub 2018 Sep 22. Curr Opin Plant Biol. 2019. PMID: 30253288 Review.
Cited by
-
Genic male and female sterility in vegetable crops.Hortic Res. 2022 Nov 19;10(1):uhac232. doi: 10.1093/hr/uhac232. eCollection 2023. Hortic Res. 2022. PMID: 36643746 Free PMC article.
-
Plant Genome Editing and the Relevance of Off-Target Changes.Plant Physiol. 2020 Aug;183(4):1453-1471. doi: 10.1104/pp.19.01194. Epub 2020 May 26. Plant Physiol. 2020. PMID: 32457089 Free PMC article.
-
Genome of 'Charleston Gray', the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the U.S. National Plant Germplasm System watermelon collection.Plant Biotechnol J. 2019 Dec;17(12):2246-2258. doi: 10.1111/pbi.13136. Epub 2019 May 7. Plant Biotechnol J. 2019. PMID: 31022325 Free PMC article.
-
Segmental duplications are hot spots of copy number variants affecting barley gene content.Plant J. 2020 Aug;103(3):1073-1088. doi: 10.1111/tpj.14784. Epub 2020 May 17. Plant J. 2020. PMID: 32338390 Free PMC article.
-
Comparison of multiple algorithms to reliably detect structural variants in pears.BMC Genomics. 2020 Jan 20;21(1):61. doi: 10.1186/s12864-020-6455-x. BMC Genomics. 2020. PMID: 31959124 Free PMC article.
References
-
- Anonymous (1973). Vademecum voor de Glastuinbouw: Glasgroenten, Glasbloemen, Champignons. (The Hague, The Netherlands: Landbouw-Economisch Instituut; ).
-
- Baker M. (2012). Structural variation: the genome’s hidden architecture. Nat. Methods 9: 133–137. - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate-a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300.
-
- Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

