Nucleomorph Genome Sequences of Two Chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata
- PMID: 26002880
- PMCID: PMC4494063
- DOI: 10.1093/gbe/evv096
Nucleomorph Genome Sequences of Two Chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata
Abstract
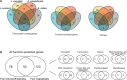
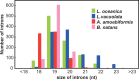
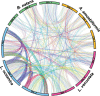
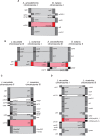
Many algal groups acquired complex plastids by the uptake of green and red algae through multiple secondary endosymbioses. As a result of gene loss and transfer during the endosymbiotic processes, algal endosymbiont nuclei disappeared in most cases. However, chlorarachniophytes and cryptophytes still possess a relict nucleus, so-called the nucleomorph, of the green and red algal endosymbiont, respectively. Nucleomorph genomes are an interesting and suitable model to study the reductive evolution of endosymbiotically derived genomes. To date, nucleomorph genomes have been sequenced in four cryptophyte species and two chlorarachniophyte species, including Bigelowiella natans (373 kb) and Lotharella oceanica (610 kb). In this study, we report complete nucleomorph genome sequences of two chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata, to gain insight into the reductive evolution of nucleomorph genomes in the chlorarachniophytes. The nucleomorph genomes consist of three chromosomes totaling 374 and 432 kb in size in A. amoebiformis and L. vacuolata, respectively. Comparative analyses among four chlorarachniophyte nucleomorph genomes revealed that these sequences share 171 function-predicted genes (86% of total 198 function-predicted nucleomorph genes), including the same set of genes encoding 17 plastid-associated proteins, and no evidence of a recent nucleomorph-to-nucleus gene transfer was found. This suggests that chlorarachniophyte nucleomorph genomes underwent most of their reductive evolution prior to the radiation of extent members of the group. However, there are slight variations in genome size, GC content, duplicated gene number, and subtelomeric regions among the four nucleomorph genomes, suggesting that the genomes might be undergoing changes that do not affect the core functions in each species.
Keywords: chlorarachniophyte; endosymbiosis; genome reduction; nucleomorph; secondary plastid.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Archibald JM. 2007. Nucleomorph genomes: structure, function, origin and evolution. Bioessays 29:392–402. - PubMed
-
- Archibald JM. 2009. The puzzle of plastid evolution. Curr Biol. 19:R81–R88. - PubMed
-
- Archibald JM, Lane CE. 2009. Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction. J Hered. 100:582–590. - PubMed
-
- Curtis BA, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492:59–65. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

