Microbiota Dynamics Associated with Environmental Conditions and Potential Roles of Cellulolytic Communities in Traditional Chinese Cereal Starter Solid-State Fermentation
- PMID: 26002897
- PMCID: PMC4495194
- DOI: 10.1128/AEM.01325-15
Microbiota Dynamics Associated with Environmental Conditions and Potential Roles of Cellulolytic Communities in Traditional Chinese Cereal Starter Solid-State Fermentation
Abstract
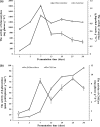
Traditional Chinese solid-state fermented cereal starters contain highly complex microbial communities and enzymes. Very little is known, however, about the microbial dynamics related to environmental conditions, and cellulolytic communities have never been proposed to exist during cereal starter fermentation. In this study, we performed Illumina MiSeq sequencing combined with PCR-denaturing gradient gel electrophoresis to investigate microbiota, coupled with clone library construction to trace cellulolytic communities in both fermentation stages. A succession of microbial assemblages was observed during the fermentation of starters. Lactobacillales and Saccharomycetales dominated the initial stages, with a continuous decline in relative abundance. However, thermotolerant and drought-resistant Bacillales, Eurotiales, and Mucorales were considerably accelerated during the heating stages, and these organisms dominated until the end of fermentation. Enterobacteriales were consistently ubiquitous throughout the process. For the cellulolytic communities, only the genera Sanguibacter, Beutenbergia, Agrobacterium, and Erwinia dominated the initial fermentation stages. In contrast, stages at high incubation temperature induced the appearance and dominance of Bacillus, Aspergillus, and Mucor. The enzymatic dynamics of amylase and glucoamylase also showed a similar trend, with the activities clearly increased in the first 7 days and subsequently decreased until the end of fermentation. Furthermore, β-glucosidase activity continuously and significantly increased during the fermentation process. Evidently, cellulolytic potential can adapt to environmental conditions by changes in the community structure during the fermentation of starters.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Bertoldo C, Antranikian G. 2002. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr Opin Microbiol 6:151–160. - PubMed
-
- Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 15:67–78. doi:10.1016/j.tifs.2003.09.004. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- SRA/PRJNA272559
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

