Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy
- PMID: 26003801
- PMCID: PMC4529488
- DOI: 10.1158/2159-8290.CD-15-0412
Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy
Abstract
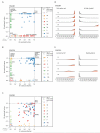
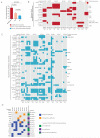
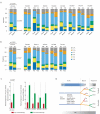
Esophageal adenocarcinomas are associated with a dismal prognosis. Deciphering the evolutionary history of this disease may shed light on therapeutically tractable targets and reveal dynamic mutational processes during the disease course and following neoadjuvant chemotherapy (NAC). We exome sequenced 40 tumor regions from 8 patients with operable esophageal adenocarcinomas, before and after platinum-containing NAC. This revealed the evolutionary genomic landscape of esophageal adenocarcinomas with the presence of heterogeneous driver mutations, parallel evolution, early genome-doubling events, and an association between high intratumor heterogeneity and poor response to NAC. Multiregion sequencing demonstrated a significant reduction in thymine to guanine mutations within a CpTpT context when comparing early and late mutational processes and the presence of a platinum signature with enrichment of cytosine to adenine mutations within a CpC context following NAC. Esophageal adenocarcinomas are characterized by early chromosomal instability leading to amplifications containing targetable oncogenes persisting through chemotherapy, providing a rationale for future therapeutic approaches.
Significance: This work illustrates dynamic mutational processes occurring during esophageal adenocarcinoma evolution and following selective pressures of platinum exposure, emphasizing the iatrogenic impact of therapy on cancer evolution. Identification of amplifications encoding targetable oncogenes maintained through NAC suggests the presence of stable vulnerabilities, unimpeded by cytotoxics, suitable for therapeutic intervention.
©2015 American Association for Cancer Research.
Figures

Comment in
-
Clonal evolution: multiregion sequencing of esophageal adenocarcinoma before and after chemotherapy.Cancer Discov. 2015 Aug;5(8):796-8. doi: 10.1158/2159-8290.CD-15-0739. Cancer Discov. 2015. PMID: 26243860
References
-
- Surveillance, Epidemiology, and End Results Program [Internet] National Cancer Institute, National Institutes of Health; Bethesda: Available from: http://seer.cancer.gov/statfacts/html/esoph.html.
-
- Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–6. - PubMed
-
- Davies AR, Gossage JA, Zylstra J, Mattsson F, Lagergren J, Maisey N, et al. Tumor stage after neoadjuvant chemotherapy determines survival after surgery for adenocarcinoma of the esophagus and esophagogastric junction. J Clin Oncol. 2014;32:2983–90. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

