Biotransformation of acetophenone and its halogen derivatives by Yarrowia lipolytica strains
- PMID: 26005401
- PMCID: PMC4438219
- DOI: 10.1007/s13213-014-0955-3
Biotransformation of acetophenone and its halogen derivatives by Yarrowia lipolytica strains
Abstract
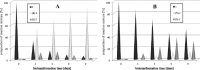
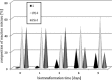
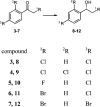
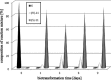
The ability of 16 strains of Yarrowia lipolytica to biotransform acetophenone and its derivatives has been studied. Thirteen of these strains were derived from a wild-type strain Y. lipolytica A-101; six had the invertase gene (SUC2) from Saccharomyces cerevisiae integrated into their genome, as well as the damaged or undamaged gene encoding orotidine-5'-phosphate decarboxylase (URA3), three had integrated the damaged URA3 gene into their genome and three were UV acetate-negative mutants, not able to growth on acetate as the sole carbon source. The other tested strains included two wild strains, A-101 and PMR-1, and an adenine auxotroph ATCC 32-338A. All strains were capable of reducing acetophenone to the R-alcohol in high enantiomeric excess (80-89 %). In all of the cultures tested, reversibility of the reduction was observed, which led to an increase in the enantiomeric excess. nantioselective reduction of the acetophenone halogen derivatives revealed that the nature and location of the halogen atom had a significant influence on the enantioselectivity of the reduction. In the culture of ATCC 32-338A, after a 3-day biotransformation of 2,4'-dibromoacetophenone the enantiopure R-alcohol was obtained at a rate of 100 % of substrate conversion. In conclusion, using these invertase-containing strains or uracyl auxotrophs provided no additional benefit in terms of biotransformation capacity over the parental strain.
Keywords: Enantiospecific reduction; Halogen derivatives of acetophenone; SUC2; URA3; Yarrowia lipolytica.
Figures

Similar articles
-
Transforming sugars into fat - lipid biosynthesis using different sugars in Yarrowia lipolytica.Yeast. 2017 Jul;34(7):293-304. doi: 10.1002/yea.3232. Epub 2017 Apr 20. Yeast. 2017. PMID: 28303649
-
Three alcohol dehydrogenase genes and one acetyl-CoA synthetase gene are responsible for ethanol utilization in Yarrowia lipolytica.Fungal Genet Biol. 2016 Oct;95:30-38. doi: 10.1016/j.fgb.2016.07.012. Epub 2016 Jul 30. Fungal Genet Biol. 2016. PMID: 27486067
-
Engineering Yarrowia lipolytica to express secretory invertase with strong FBA1IN promoter.Yeast. 2012 Feb;29(2):59-72. doi: 10.1002/yea.1917. Epub 2011 Dec 29. Yeast. 2012. PMID: 22222800
-
Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker.Curr Genet. 1989 Oct;16(4):253-60. doi: 10.1007/BF00422111. Curr Genet. 1989. PMID: 2627713
-
Bioconversion of R-(+)-limonene to perillic acid by the yeast Yarrowia lipolytica.Braz J Microbiol. 2014 Mar 10;44(4):1075-80. doi: 10.1590/S1517-83822014005000008. eCollection 2013 Dec. Braz J Microbiol. 2014. PMID: 24688495 Free PMC article.
Cited by
-
Haloferax volcanii for biotechnology applications: challenges, current state and perspectives.Appl Microbiol Biotechnol. 2020 Feb;104(4):1371-1382. doi: 10.1007/s00253-019-10314-2. Epub 2019 Dec 20. Appl Microbiol Biotechnol. 2020. PMID: 31863144 Free PMC article. Review.
-
Highly enantioselective production of (R)-halohydrins with whole cells of Rhodotorula rubra KCh 82 culture.Int J Mol Sci. 2014 Dec 4;15(12):22392-404. doi: 10.3390/ijms151222392. Int J Mol Sci. 2014. PMID: 25486054 Free PMC article.
-
Haloferax volcanii as immobilised whole cell biocatalyst: new applications for halophilic systems.Appl Microbiol Biotechnol. 2019 May;103(9):3807-3817. doi: 10.1007/s00253-019-09725-y. Epub 2019 Mar 15. Appl Microbiol Biotechnol. 2019. PMID: 30877354 Free PMC article.
References
-
- Basavaiah D, Reddy GJ, Chandrashekar V. A new chiral catalytic source with an N–P = O structural framework containing a proximal hydroxyl group for the borane-mediated asymmetric reduction of prochiral ketones. Tetrahedron Asymmetry. 2004;15:47–52. doi: 10.1016/j.tetasy.2003.11.002. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
