Multidimensional metrics for estimating phage abundance, distribution, gene density, and sequence coverage in metagenomes
- PMID: 26005436
- PMCID: PMC4424905
- DOI: 10.3389/fmicb.2015.00381
Multidimensional metrics for estimating phage abundance, distribution, gene density, and sequence coverage in metagenomes
Abstract
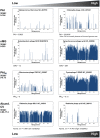
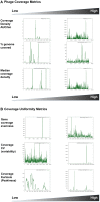
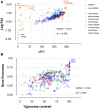
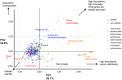
Phages are the most abundant biological entities on Earth and play major ecological roles, yet the current sequenced phage genomes do not adequately represent their diversity, and little is known about the abundance and distribution of these sequenced genomes in nature. Although the study of phage ecology has benefited tremendously from the emergence of metagenomic sequencing, a systematic survey of phage genes and genomes in various ecosystems is still lacking, and fundamental questions about phage biology, lifestyle, and ecology remain unanswered. To address these questions and improve comparative analysis of phages in different metagenomes, we screened a core set of publicly available metagenomic samples for sequences related to completely sequenced phages using the web tool, Phage Eco-Locator. We then adopted and deployed an array of mathematical and statistical metrics for a multidimensional estimation of the abundance and distribution of phage genes and genomes in various ecosystems. Experiments using those metrics individually showed their usefulness in emphasizing the pervasive, yet uneven, distribution of known phage sequences in environmental metagenomes. Using these metrics in combination allowed us to resolve phage genomes into clusters that correlated with their genotypes and taxonomic classes as well as their ecological properties. We propose adding this set of metrics to current metaviromic analysis pipelines, where they can provide insight regarding phage mosaicism, habitat specificity, and evolution.
Keywords: bacteriophage; ecology; genomics; metagenomics; virus.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

