Diversity of acid stress resistant variants of Listeria monocytogenes and the potential role of ribosomal protein S21 encoded by rpsU
- PMID: 26005439
- PMCID: PMC4424878
- DOI: 10.3389/fmicb.2015.00422
Diversity of acid stress resistant variants of Listeria monocytogenes and the potential role of ribosomal protein S21 encoded by rpsU
Abstract
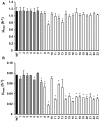
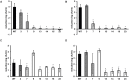
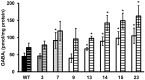
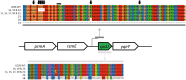
The dynamic response of microorganisms to environmental conditions depends on the behavior of individual cells within the population. Adverse environments can select for stable stress resistant subpopulations. In this study, we aimed to get more insight in the diversity within Listeria monocytogenes LO28 populations, and the genetic basis for the increased resistance of stable resistant fractions isolated after acid exposure. Phenotypic cluster analysis of 23 variants resulted in three clusters and four individual variants and revealed multiple-stress resistance, with both unique and overlapping features related to stress resistance, growth, motility, biofilm formation, and virulence indicators. A higher glutamate decarboxylase activity correlated with increased acid resistance. Whole genome sequencing revealed mutations in rpsU, encoding ribosomal protein S21 in the largest phenotypic cluster, while mutations in ctsR, which were previously shown to be responsible for increased resistance of heat and high hydrostatic pressure resistant variants, were not found in the acid resistant variants. This underlined that large population diversity exists within one L. monocytogenes strain and that different adverse conditions drive selection for different variants. The finding that acid stress selects for rpsU variants provides potential insights in the mechanisms underlying population diversity of L. monocytogenes.
Keywords: SNP analysis; glutamate decarboxylase; heterogeneity; phenotypic characterization; trade-off; whole genome sequencing.
Figures
Similar articles
-
Amino acid substitutions in ribosomal protein RpsU enable switching between high fitness and multiple-stress resistance in Listeria monocytogenes.Int J Food Microbiol. 2021 Aug 2;351:109269. doi: 10.1016/j.ijfoodmicro.2021.109269. Epub 2021 Jun 1. Int J Food Microbiol. 2021. PMID: 34102570
-
Population diversity of Listeria monocytogenes LO28: phenotypic and genotypic characterization of variants resistant to high hydrostatic pressure.Appl Environ Microbiol. 2010 Apr;76(7):2225-33. doi: 10.1128/AEM.02434-09. Epub 2010 Feb 5. Appl Environ Microbiol. 2010. PMID: 20139309 Free PMC article.
-
Gene profiling-based phenotyping for identification of cellular parameters that contribute to fitness, stress-tolerance and virulence of Listeria monocytogenes variants.Int J Food Microbiol. 2018 Oct 20;283:14-21. doi: 10.1016/j.ijfoodmicro.2018.06.003. Epub 2018 Jun 7. Int J Food Microbiol. 2018. PMID: 29935377
-
Impact of Pathogen Population Heterogeneity and Stress-Resistant Variants on Food Safety.Annu Rev Food Sci Technol. 2016;7:439-56. doi: 10.1146/annurev-food-041715-033128. Epub 2016 Jan 11. Annu Rev Food Sci Technol. 2016. PMID: 26772414 Review.
-
Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis.Appl Microbiol Biotechnol. 2018 Apr;102(8):3475-3485. doi: 10.1007/s00253-018-8852-5. Epub 2018 Mar 2. Appl Microbiol Biotechnol. 2018. PMID: 29500754 Review.
Cited by
-
Ribosomal mutations enable a switch between high fitness and high stress resistance in Listeria monocytogenes.Front Microbiol. 2024 Mar 28;15:1355268. doi: 10.3389/fmicb.2024.1355268. eCollection 2024. Front Microbiol. 2024. PMID: 38605704 Free PMC article.
-
Ribosomes lacking bS21 gain function to regulate protein synthesis in Flavobacterium johnsoniae.Nucleic Acids Res. 2023 Feb 28;51(4):1927-1942. doi: 10.1093/nar/gkad047. Nucleic Acids Res. 2023. PMID: 36727479 Free PMC article.
-
Identification of Novel Genes Mediating Survival of Salmonella on Low-Moisture Foods via Transposon Sequencing Analysis.Front Microbiol. 2020 May 15;11:726. doi: 10.3389/fmicb.2020.00726. eCollection 2020. Front Microbiol. 2020. PMID: 32499760 Free PMC article.
-
Different Transcriptional Responses from Slow and Fast Growth Rate Strains of Listeria monocytogenes Adapted to Low Temperature.Front Microbiol. 2016 Mar 1;7:229. doi: 10.3389/fmicb.2016.00229. eCollection 2016. Front Microbiol. 2016. PMID: 26973610 Free PMC article.
-
Genotypes Associated with Listeria monocytogenes Isolates Displaying Impaired or Enhanced Tolerances to Cold, Salt, Acid, or Desiccation Stress.Front Microbiol. 2017 Mar 8;8:369. doi: 10.3389/fmicb.2017.00369. eCollection 2017. Front Microbiol. 2017. PMID: 28337186 Free PMC article.
References
-
- Akanuma G., Nanamiya H., Natori Y., Yano K., Suzuki S., Omata S., et al. (2012). Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J. Bacteriol. 194 6282–6291 10.1128/JB.01544-12 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

