The schizophrenia risk gene product miR-137 alters presynaptic plasticity
- PMID: 26005852
- PMCID: PMC4506960
- DOI: 10.1038/nn.4023
The schizophrenia risk gene product miR-137 alters presynaptic plasticity
Erratum in
-
Addendum: The schizophrenia risk gene product miR-137 alters presynaptic plasticity.Nat Neurosci. 2016 Jul 26;19(8):1115. doi: 10.1038/nn0816-1115c. Nat Neurosci. 2016. PMID: 27459408 No abstract available.
Abstract
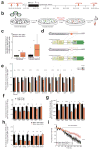
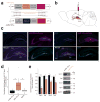
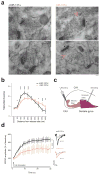
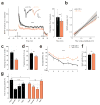
Noncoding variants in the human MIR137 gene locus increase schizophrenia risk with genome-wide significance. However, the functional consequence of these risk alleles is unknown. Here we examined induced human neurons harboring the minor alleles of four disease-associated single nucleotide polymorphisms in MIR137. We observed increased MIR137 levels compared to those in major allele-carrying cells. microRNA-137 gain of function caused downregulation of the presynaptic target genes complexin-1 (Cplx1), Nsf and synaptotagmin-1 (Syt1), leading to impaired vesicle release. In vivo, miR-137 gain of function resulted in changes in synaptic vesicle pool distribution, impaired induction of mossy fiber long-term potentiation and deficits in hippocampus-dependent learning and memory. By sequestering endogenous miR-137, we were able to ameliorate the synaptic phenotypes. Moreover, reinstatement of Syt1 expression partially restored synaptic plasticity, demonstrating the importance of Syt1 as a miR-137 target. Our data provide new insight into the mechanism by which miR-137 dysregulation can impair synaptic plasticity in the hippocampus.
Figures
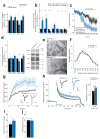
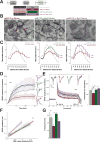
Comment in
-
Synaptic plasticity: Micro-level disruption.Nat Rev Neurosci. 2015 Jul;16(7):373. doi: 10.1038/nrn3989. Nat Rev Neurosci. 2015. PMID: 26087678 No abstract available.
-
MIR137: big impacts from small changes.Nat Neurosci. 2015 Jul;18(7):931-3. doi: 10.1038/nn.4045. Nat Neurosci. 2015. PMID: 26108719 No abstract available.
Similar articles
-
MIR137: big impacts from small changes.Nat Neurosci. 2015 Jul;18(7):931-3. doi: 10.1038/nn.4045. Nat Neurosci. 2015. PMID: 26108719 No abstract available.
-
Synaptic plasticity: Micro-level disruption.Nat Rev Neurosci. 2015 Jul;16(7):373. doi: 10.1038/nrn3989. Nat Rev Neurosci. 2015. PMID: 26087678 No abstract available.
-
The N-ethylmaleimide-sensitive factor and dysbindin interact to modulate synaptic plasticity.J Neurosci. 2015 May 13;35(19):7643-53. doi: 10.1523/JNEUROSCI.4724-14.2015. J Neurosci. 2015. PMID: 25972187 Free PMC article.
-
RIM1: an edge for presynaptic plasticity.Trends Neurosci. 2002 Jul;25(7):329-32. doi: 10.1016/s0166-2236(02)02193-8. Trends Neurosci. 2002. PMID: 12079752 Review.
-
Active zones for presynaptic plasticity in the brain.Mol Psychiatry. 2005 Feb;10(2):185-200; image 131. doi: 10.1038/sj.mp.4001628. Mol Psychiatry. 2005. PMID: 15630409 Review.
Cited by
-
MicroRNA-137 Inhibits EFNB2 Expression Affected by a Genetic Variant and Is Expressed Aberrantly in Peripheral Blood of Schizophrenia Patients.EBioMedicine. 2016 Oct;12:133-142. doi: 10.1016/j.ebiom.2016.09.012. Epub 2016 Sep 15. EBioMedicine. 2016. PMID: 27650867 Free PMC article.
-
The Neurotoxic Effect of Environmental Temperature Variation in Adult Zebrafish (Danio rerio).Int J Mol Sci. 2023 Oct 29;24(21):15735. doi: 10.3390/ijms242115735. Int J Mol Sci. 2023. PMID: 37958719 Free PMC article.
-
MicroRNA-29 is an essential regulator of brain maturation through regulation of CH methylation.Cell Rep. 2021 Apr 6;35(1):108946. doi: 10.1016/j.celrep.2021.108946. Cell Rep. 2021. PMID: 33826889 Free PMC article.
-
miR-137 regulates PTP61F, affecting insulin signaling, metabolic homeostasis, and starvation resistance in Drosophila.Proc Natl Acad Sci U S A. 2024 Jan 30;121(5):e2319475121. doi: 10.1073/pnas.2319475121. Epub 2024 Jan 22. Proc Natl Acad Sci U S A. 2024. PMID: 38252824 Free PMC article.
-
Genomics and the future of psychopharmacology: MicroRNAs offer novel therapeutics .Dialogues Clin Neurosci. 2019;21(2):131-148. doi: 10.31887/DCNS.2019.21.2/dgurwitz. Dialogues Clin Neurosci. 2019. PMID: 31636487 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

