Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus
- PMID: 26006014
- PMCID: PMC4512288
- DOI: 10.1038/ni.3175
Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus
Abstract
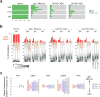
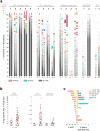
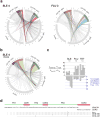
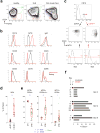
Acute systemic lupus erythematosus (SLE) courses with surges of antibody-secreting cells (ASCs) whose origin, diversity and contribution to serum autoantibodies remain unknown. Here, deep sequencing, proteomic profiling of autoantibodies and single-cell analysis demonstrated highly diversified ASCs punctuated by clones expressing the variable heavy-chain region VH4-34 that produced dominant serum autoantibodies. A fraction of ASC clones contained autoantibodies without mutation, a finding consistent with differentiation outside the germinal centers. A substantial ASC segment was derived from a distinct subset of newly activated naive cells of considerable clonality that persisted in the circulation for several months. Thus, selection of SLE autoreactivities occurred during polyclonal activation, with prolonged recruitment of recently activated naive B cells. Our findings shed light on the pathogenesis of SLE, help explain the benefit of agents that target B cells and should facilitate the design of future therapies.
Figures




Comment in
-
How many memories does it take to make an SLE flare?Nat Immunol. 2015 Jul;16(7):685-7. doi: 10.1038/ni.3209. Nat Immunol. 2015. PMID: 26086136 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

