The deubiquitinase ataxin-3 requires Rad23 and DnaJ-1 for its neuroprotective role in Drosophila melanogaster
- PMID: 26007638
- PMCID: PMC4710962
- DOI: 10.1016/j.nbd.2015.05.010
The deubiquitinase ataxin-3 requires Rad23 and DnaJ-1 for its neuroprotective role in Drosophila melanogaster
Abstract
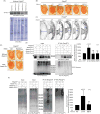
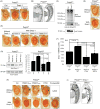
Ataxin-3 is a deubiquitinase and polyglutamine (polyQ) disease protein with a protective role in Drosophila melanogaster models of neurodegeneration. In the fruit fly, wild-type ataxin-3 suppresses toxicity from several polyQ disease proteins, including a pathogenic version of itself that causes spinocerebellar ataxia type 3 and pathogenic huntingtin, which causes Huntington's disease. The molecular partners of ataxin-3 in this protective function are unclear. Here, we report that ataxin-3 requires its direct interaction with the ubiquitin-binding and proteasome-associated protein, Rad23 (known as hHR23A/B in mammals) in order to suppress toxicity from polyQ species in Drosophila. According to additional studies, ataxin-3 does not rely on autophagy or the proteasome to suppress polyQ-dependent toxicity in fly eyes. Instead this deubiquitinase, through its interaction with Rad23, leads to increased protein levels of the co-chaperone DnaJ-1 and depends on it to protect against degeneration. Through DnaJ-1, our data connect ataxin-3 and Rad23 to protective processes involved with protein folding rather than increased turnover of toxic polyQ species.
Keywords: Ataxin-3; Chaperone; Deubiquitinase; Drosophila; Machado–Joseph disease; Polyglutamine; Ubiquitin.
Copyright © 2015. Published by Elsevier Inc.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Becker J, et al. Hydrogen peroxide activates immediate binding of a Drosophila factor to DNA heat-shock regulatory element in vivo and in vitro. Eur J Biochem. 1990;189:553–558. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

