Architectural proteins, transcription, and the three-dimensional organization of the genome
- PMID: 26008126
- PMCID: PMC4598269
- DOI: 10.1016/j.febslet.2015.05.025
Architectural proteins, transcription, and the three-dimensional organization of the genome
Abstract
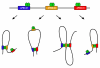
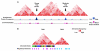
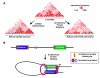
Architectural proteins mediate interactions between distant sequences in the genome. Two well-characterized functions of architectural protein interactions include the tethering of enhancers to promoters and bringing together Polycomb-containing sites to facilitate silencing. The nature of which sequences interact genome-wide appears to be determined by the orientation of the architectural protein binding sites as well as the number and identity of architectural proteins present. Ultimately, long range chromatin interactions result in the formation of loops within the chromatin fiber. In this review, we discuss data suggesting that architectural proteins mediate long range chromatin interactions that both facilitate and hinder neighboring interactions, compartmentalizing the genome into regions of highly interacting chromatin domains.
Keywords: CCCTC-binding factor; Chromatin; Epigenetics; Insulator; Topologically associating domain.
Copyright © 2015 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nature reviews Molecular cell biology. 2015;16:245–57. - PubMed
-
- Lupiáñez Darío G., Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz John M., Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas Stefan A., Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell - PMC - PubMed
-
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. - PMC - PubMed
-
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

