Pitfalls in histone propionylation during bottom-up mass spectrometry analysis
- PMID: 26010583
- PMCID: PMC5032999
- DOI: 10.1002/pmic.201400569
Pitfalls in histone propionylation during bottom-up mass spectrometry analysis
Abstract
Despite their important role in regulating gene expression, posttranslational histone modifications remain technically challenging to analyze. For identification by bottom-up MS, propionylation is required prior to and following trypsin digestion. Hereby, more hydrophobic peptides are generated enabling RP HPLC separation. When histone dynamics are studied in a quantitative manner, specificity, and efficiency of this chemical derivatization are crucial. Therefore we examined eight different protocols, including two different propionylation reagents. This revealed amidation (up to 70%) and methylation (up to 9%) of carboxyl groups as a side reaction. Moreover, incomplete (up to 85%) as well as a specific propionylation (up to 63%) can occur, depending on the protocol. These results highlight the possible pitfalls and implications for data analysis when doing bottom-up MS on histones.
Keywords: Cell biology; Histone; MS-LC-MS; Method optimization; Propionylation.
© 2015 The Authors. PROTEOMICS Published by Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
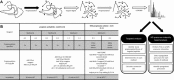
 ) as well as postdigestion (
) as well as postdigestion ( ), followed by LCMSMS analysis. During the first propionylation reaction K, monomethylated K and the protein N‐terminus become derivatized. After digestion the newly generated peptide N‐termini get propionylated as well. The generated LCMSMS data were analyzed using two approaches: (i) left box: a targeted approach, defining the conversion rate based on identified peptides (ii) right box: an untargeted approach, based on differential MS precursor intensities in between methods. The first strategy can be used to determine efficiency and specificity of the protocol, the latter is used to monitor for unexpected side reactions that differ in between protocols. (B) Table representing the differences between the propionylation methods. The four different methods vary in propionylation reagent, buffer, and reaction temperature. Methods A to C are performed with a single (1×) as well as a double (2×) round of propionylation before and after digestion, marked as Method X1 and X2, respectively. Method D is carried out with a 150 times molar excess (Method D1) and a 600 × molar excess (Method D2) of NHS‐propionate to the bovine histones.
), followed by LCMSMS analysis. During the first propionylation reaction K, monomethylated K and the protein N‐terminus become derivatized. After digestion the newly generated peptide N‐termini get propionylated as well. The generated LCMSMS data were analyzed using two approaches: (i) left box: a targeted approach, defining the conversion rate based on identified peptides (ii) right box: an untargeted approach, based on differential MS precursor intensities in between methods. The first strategy can be used to determine efficiency and specificity of the protocol, the latter is used to monitor for unexpected side reactions that differ in between protocols. (B) Table representing the differences between the propionylation methods. The four different methods vary in propionylation reagent, buffer, and reaction temperature. Methods A to C are performed with a single (1×) as well as a double (2×) round of propionylation before and after digestion, marked as Method X1 and X2, respectively. Method D is carried out with a 150 times molar excess (Method D1) and a 600 × molar excess (Method D2) of NHS‐propionate to the bovine histones.
 ), desired products (
), desired products ( ), overpropionylated products (
), overpropionylated products ( ). This clearly illustrates the increasing retention that is induced by propionylation. (C) Radar chart representing the average conversion rate for eight targeted peptides. Each peptide is located on one angle of the radar chart and each method is represented by another color. The conversion rate for each peptide using the different methods is shown on the radius, whereby a conversion rate of 0 is located in the center, increasing outwards.
). This clearly illustrates the increasing retention that is induced by propionylation. (C) Radar chart representing the average conversion rate for eight targeted peptides. Each peptide is located on one angle of the radar chart and each method is represented by another color. The conversion rate for each peptide using the different methods is shown on the radius, whereby a conversion rate of 0 is located in the center, increasing outwards.
Comment in
-
Properly reading the histone code by MS-based proteomics.Proteomics. 2015 Sep;15(17):2901-2. doi: 10.1002/pmic.201500298. Proteomics. 2015. PMID: 26223514
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

