Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production
- PMID: 26011187
- PMCID: PMC11389102
- DOI: 10.1111/pbi.12403
Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production
Abstract
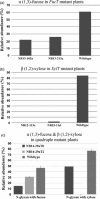
Biopharmaceutical glycoproteins produced in plants carry N-glycans with plant-specific residues core α(1,3)-fucose and β(1,2)-xylose, which can significantly impact the activity, stability and immunogenicity of biopharmaceuticals. In this study, we have employed sequence-specific transcription activator-like effector nucleases (TALENs) to knock out two α(1,3)-fucosyltransferase (FucT) and the two β(1,2)-xylosyltransferase (XylT) genes within Nicotiana benthamiana to generate plants with improved capacity to produce glycoproteins devoid of plant-specific residues. Among plants regenerated from N. benthamiana protoplasts transformed with TALENs targeting either the FucT or XylT genes, 50% (80 of 160) and 73% (94 of 129) had mutations in at least one FucT or XylT allele, respectively. Among plants regenerated from protoplasts transformed with both TALEN pairs, 17% (18 of 105) had mutations in all four gene targets, and 3% (3 of 105) plants had mutations in all eight alleles comprising both gene families; these mutations were transmitted to the next generation. Endogenous proteins expressed in the complete knockout line had N-glycans that lacked β(1,2)-xylose and had a significant reduction in core α(1,3)-fucose levels (40% of wild type). A similar phenotype was observed in the N-glycans of a recombinant rituximab antibody transiently expressed in the homozygous mutant plants. More importantly, the most desirable glycoform, one lacking both core α(1,3)-fucose and β(1,2)-xylose residues, increased in the antibody from 2% when produced in the wild-type line to 55% in the mutant line. These results demonstrate the power of TALENs for multiplexed gene editing. Furthermore, the mutant N. benthamiana lines provide a valuable platform for producing highly potent biopharmaceutical products.
Keywords: Gene editing; glyco-engineering; plant-derived pharmaceuticals; transcription activator-like effector nucleases.
© 2015 Society for Experimental Biology, Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
JL, TJS, ZLD, SL, BMC, EER, AC, AD, AY, AR, LM, DFV and FZ are employees of Cellectis Plant Sciences Inc., a subsidiary of Cellectis SA. FC is an employee of Cellectis SA. POL and MAD are employees of Medicago R&D, a subsidiary of Medicago Inc.
Figures



References
-
- Arnould, S. , Chames, P. , Perez, C. , Lacroix, E. , Duclert, A. , Epinat, J.C. , Stricher, F. , Petit, A.S. , Patin, A. , Guillier, S. , Rolland, S. , Prieto, J. , Blanco, F.J. , Bravo, J. , Montoya, G. , Serrano, L. , Duchateau, P. and Paques, F. (2006) Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 355, 443–458. - PubMed
-
- Baltes, N.J. and Voytas, D.F. (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33, 120–131. - PubMed
-
- Bombarely, A. , Rosli, H.G. , Vrebalov, J. , Moffett, P. , Mueller, L.A. and Martin, G.B. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant‐microbe biology research. Mol. Plant‐Microbe Interact. 25, 1523–1530. - PubMed
-
- Cabanes‐Macheteau, M. , Fitchette‐Laine, A.C. , Loutelier‐Bourhis, C. , Lange, C. , Vine, N.D. , Ma, J.K. , Lerouge, P. and Faye, L. (1999) N‐Glycosylation of a mouse IgG expressed in transgenic tobacco plants. Glycobiology, 9, 365–372. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

