Full UPF3B function is critical for neuronal differentiation of neural stem cells
- PMID: 26012578
- PMCID: PMC4445987
- DOI: 10.1186/s13041-015-0122-1
Full UPF3B function is critical for neuronal differentiation of neural stem cells
Abstract
Background: Mutation in the UPF3B gene on chromosome X is implicated in neurodevelopmental disorders including X-linked intellectual disability, autism and schizophrenia. The protein UPF3B is involved in the nonsense-mediated mRNA decay pathway (NMD) that controls mRNA stability and functions in the prevention of the synthesis of truncated proteins.
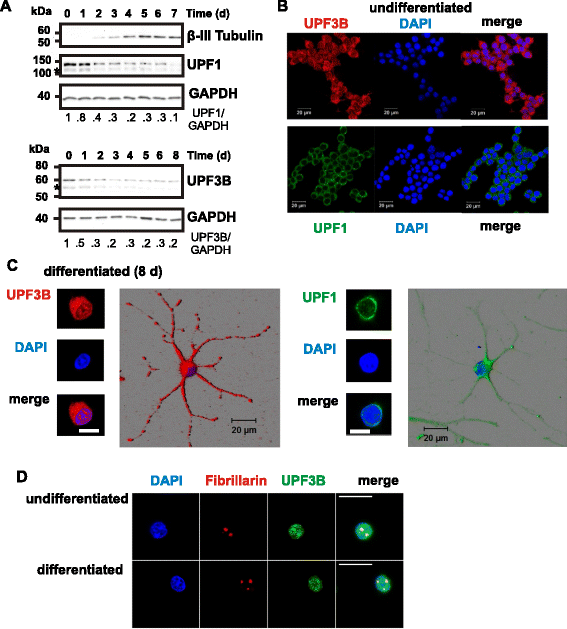
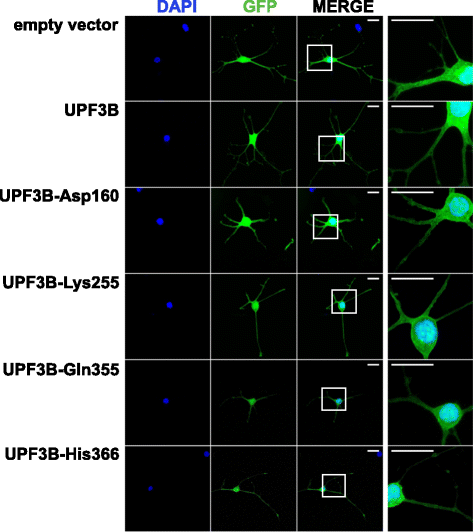
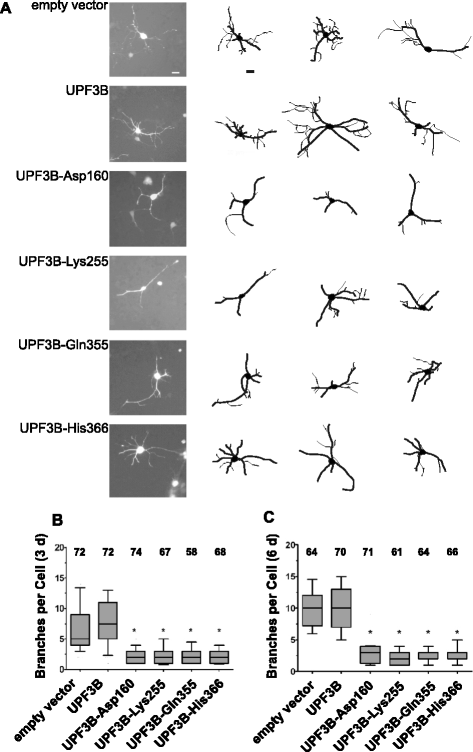
Results: Here we show that NMD pathway components UPF3B and UPF1 are down-regulated during differentiation of neural stem cells into neurons. Using tethered function assays we found that UPF3B missense mutations described in families with neurodevelopmental disorders reduced the activity of UPF3B protein in NMD. In neural stem cells, UPF3B protein was detected in the cytoplasm and in the nucleus. Similarly in neurons, UPF3B protein was detected in neurites, the somatic cytoplasm and in the nucleus. In both cell types nuclear UPF3B protein was enriched in the nucleolus. Using GFP tagged UPF3B proteins we found that the missense mutations did not affect the cellular localisation. Expression of missense mutant UPF3B disturbed neuronal differentiation and reduced the complexity of the branching of neurites. Neuronal differentiation was similarly affected in the presence of the NMD inhibitor Amlexanox. The expression of mutant UPF3B proteins lead to a subtle increase in mRNA levels of selected NMD targets.
Conclusions: Together our findings indicate that, despite the down-regulation of NMD factors, functional NMD is critical for neuronal differentiation. We propose that the neurodevelopmental phenotype of UPF3B missense mutation is caused by impairment of NMD function altering neuronal differentiation.
Figures



References
-
- Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry. 2010;15:767–776. doi: 10.1038/mp.2009.14. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

