Isolation and Characterization of a Thionin Proprotein-processing Enzyme from Barley
- PMID: 26013828
- PMCID: PMC4505051
- DOI: 10.1074/jbc.M115.647859
Isolation and Characterization of a Thionin Proprotein-processing Enzyme from Barley
Abstract
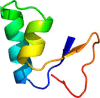
Thionins are plant-specific antimicrobial peptides that have been isolated from the endosperm and leaves of cereals, from the leaves of mistletoes, and from several other plant species. They are generally basic peptides with three or four disulfide bridges and a molecular mass of ~5 kDa. Thionins are produced as preproproteins consisting of a signal peptide, the thionin domain, and an acidic domain. Previously, only mature thionin peptides have been isolated from plants, and in addition to removal of the signal peptide, at least one cleavage processing step between the thionin and the acidic domain is necessary to release the mature thionin. In this work, we identified a thionin proprotein-processing enzyme (TPPE) from barley. Purification of the enzyme was guided by an assay that used a quenched fluorogenic peptide comprising the amino acid sequence between the thionin and the acidic domain of barley leaf-specific thionin. The barley TPPE was identified as a serine protease (BAJ93208) and expressed in Escherichia coli as a strep tag-labeled protein. The barley BTH6 thionin proprotein was produced in E. coli using the vector pETtrx1a and used as a substrate. We isolated and sequenced the BTH6 thionin from barley to confirm the N and C terminus of the peptide in planta. Using an in vitro enzymatic assay, the recombinant TPPE was able to process the quenched fluorogenic peptide and to cleave the acidic domain at least at six sites releasing the mature thionin from the proprotein. Moreover, it was found that the intrinsic three-dimensional structure of the BTH6 thionin domain prevents cleavage of the mature BTH6 thionin by the TPPE.
Keywords: antimicrobial peptide; plant defense; proprotein; proprotein convertase subtilisin/kexin type 9 (PCSK9); recombinant protein expression; serine protease; subtilase; thionin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Benko-Iseppon A. M., Galdino S. L., Calsa T., Jr., Kido E. A., Tossi A., Belarmino L. C., Crovella S. (2010) Overview on plant antimicrobial peptides. Curr. Protein Pept. Sci. 11, 181–188 - PubMed
-
- Padovan L., Scocchi M., Tossi A. (2010) Structural aspects of plant antimicrobial peptides. Curr. Protein Pept. Sci. 11, 210–219 - PubMed
-
- Balls A. K., Hale W. S., Harris T. H. (1942) A crystalline protein obtained from a lipoprotein of wheat flour. Cereal Chem. 19, 279–288
-
- Fisher N., Redman D. G., Elton G. A. (1968) Fractionation and characterization of purothionin. Cereal Chem. 45, 48–57
-
- Bohlman H., Apel K. (1991) Thionins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 227–240
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

