Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics
- PMID: 26014286
- PMCID: PMC4444815
- DOI: 10.1038/srep09833
Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics
Abstract
Outbreaks in fish of motile Aeromonad septicemia (MAS) caused by Aeromonas hydrophila have caused a great concern worldwide. Here, for the first time, we provide two complete genomes of epidemic A. hydrophila strains isolated in China. To gain an insight into the pathogenicity of epidemic A. hydrophila, we performed comparative genomic analyses of five epidemic strains belonging to sequence type (ST) 251, together with the environmental strain ATCC 7966(T). We found that the known virulence factors, including a type III secretion system, a type VI secretion system and lateral flagella, are not required for the high virulence of the ST251 clonal group. Additionally, our work identifies three utilization pathways for myo-inositol, sialic acid and L-fucose providing clues regarding the factors that underlie the epidemic and virulent nature of ST251 A. hydrophila. Based on the geographical distribution and biological resources of the ST251 clonal group, we conclude that ST251 is a high-risk clonal group of A. hydrophila which may be responsible for the MAS outbreaks in China and the southeastern United States.
Figures
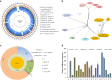

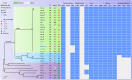
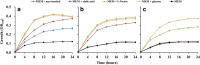
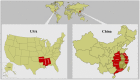
References
-
- Zhang X., Yang W., Wu H., Gong X. & Li A. Multilocus sequence typing revealed a clonal lineage of Aeromonas hydrophila caused motile Aeromonas septicemia outbreaks in pond-cultured cyprinid fish in an epidemic area in central China. Aquaculture 432, 1–6 (2014).
-
- Pridgeon J. W. & Klesius P. H. Molecular identification and virulence of three Aeromonas hydrophila isolates cultured from infected channel catfish during a disease outbreak in west Alabama (USA) in 2009. Dis. Aquat. Organ 94, 249–53 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

