Centrin-2 (Cetn2) mediated regulation of FGF/FGFR gene expression in Xenopus
- PMID: 26014913
- PMCID: PMC4650658
- DOI: 10.1038/srep10283
Centrin-2 (Cetn2) mediated regulation of FGF/FGFR gene expression in Xenopus
Abstract
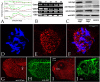
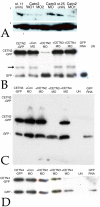
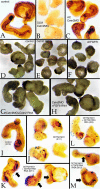
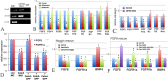
Centrins (Cetns) are highly conserved, widely expressed, and multifunctional Ca(2+)-binding eukaryotic signature proteins best known for their roles in ciliogenesis and as critical components of the global genome nucleotide excision repair system. Two distinct Cetn subtypes, Cetn2-like and Cetn3-like, have been recognized and implicated in a range of cellular processes. In the course of morpholino-based loss of function studies in Xenopus laevis, we have identified a previously unreported Cetn2-specific function, namely in fibroblast growth factor (FGF) mediated signaling, specifically through the regulation of FGF and FGF receptor RNA levels. Cetn2 was found associated with the RNA polymerase II binding sites of the Cetn2-regulated FGF8 and FGFR1a genes, but not at the promoter of a gene (BMP4) whose expression was altered indirectly in Cent2 morphant embryos. These observations point to a previously unexpected role of Cetn2 in the regulation of gene expression and embryonic development.
Figures
Similar articles
-
Temporal and spatial expression of FGF ligands and receptors during Xenopus development.Dev Dyn. 2009 Jun;238(6):1467-79. doi: 10.1002/dvdy.21913. Dev Dyn. 2009. PMID: 19322767 Free PMC article.
-
Docking protein SNT1 is a critical mediator of fibroblast growth factor signaling during Xenopus embryonic development.Dev Dyn. 2002 Mar;223(2):216-28. doi: 10.1002/dvdy.10048. Dev Dyn. 2002. PMID: 11836786
-
Genomic organization and modulation of gene expression of the TGF-β and FGF pathways in the allotetraploid frog Xenopus laevis.Dev Biol. 2017 Jun 15;426(2):336-359. doi: 10.1016/j.ydbio.2016.09.016. Epub 2016 Sep 28. Dev Biol. 2017. PMID: 27692744
-
Transgenic frogs and FGF signalling in early development.Trends Genet. 1996 Nov;12(11):439-40. doi: 10.1016/0168-9525(96)30105-4. Trends Genet. 1996. PMID: 8973142 Review. No abstract available.
-
The role of FGF signaling in guiding coordinate movement of cell groups: guidance cue and cell adhesion regulator?Cell Adh Migr. 2012 Sep-Oct;6(5):397-403. doi: 10.4161/cam.21103. Epub 2012 Sep 1. Cell Adh Migr. 2012. PMID: 23076054 Free PMC article. Review.
Cited by
-
Ciliary IFT80 regulates dental pulp stem cells differentiation by FGF/FGFR1 and Hh/BMP2 signaling.Int J Biol Sci. 2019 Aug 6;15(10):2087-2099. doi: 10.7150/ijbs.27231. eCollection 2019. Int J Biol Sci. 2019. PMID: 31592124 Free PMC article.
-
Tetrahymena Poc5 is a transient basal body component that is important for basal body maturation.J Cell Sci. 2020 Jun 4;133(11):jcs240838. doi: 10.1242/jcs.240838. J Cell Sci. 2020. PMID: 32350068 Free PMC article.
-
Centrin 3 is an inhibitor of centrosomal Mps1 and antagonizes centrin 2 function.Mol Biol Cell. 2015 Nov 1;26(21):3741-53. doi: 10.1091/mbc.E14-07-1248. Epub 2015 Sep 9. Mol Biol Cell. 2015. PMID: 26354417 Free PMC article.
-
Targeting the Interplay between HDACs and DNA Damage Repair for Myeloma Therapy.Int J Mol Sci. 2021 Sep 27;22(19):10406. doi: 10.3390/ijms221910406. Int J Mol Sci. 2021. PMID: 34638744 Free PMC article. Review.
-
Identifying domains of EFHC1 involved in ciliary localization, ciliogenesis, and the regulation of Wnt signaling.Dev Biol. 2016 Mar 15;411(2):257-265. doi: 10.1016/j.ydbio.2016.01.004. Epub 2016 Jan 16. Dev Biol. 2016. PMID: 26783883 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

