An Optimized Informatics Pipeline for Mass Spectrometry-Based Peptidomics
- PMID: 26015166
- PMCID: PMC4655184
- DOI: 10.1007/s13361-015-1169-z
An Optimized Informatics Pipeline for Mass Spectrometry-Based Peptidomics
Abstract
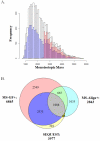
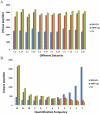
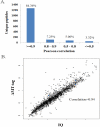
The comprehensive MS analysis of the peptidome, the intracellular and intercellular products of protein degradation, has the potential to provide novel insights on endogenous proteolytic processing and its utility in disease diagnosis and prognosis. Along with the advances in MS instrumentation and related platforms, a plethora of proteomics data analysis tools have been applied for direct use in peptidomics; however, an evaluation of the currently available informatics pipelines for peptidomics data analysis has yet to be reported. In this study, we began by evaluating the results of several popular MS/MS database search engines, including MS-GF+, SEQUEST, and MS-Align+, for peptidomics data analysis, followed by identification and label-free quantification using the well-established accurate mass and time (AMT) tag and newly developed informed quantification (IQ) approaches, both based on direct LC-MS analysis. Our results demonstrated that MS-GF+ outperformed both SEQUEST and MS-Align+ in identifying peptidome peptides. Using a database established from MS-GF+ peptide identifications, both the AMT tag and IQ approaches provided significantly deeper peptidome coverage and less missing data for each individual data set than the MS/MS methods, while achieving robust label-free quantification. Besides having an excellent correlation with the AMT tag quantification results, IQ also provided slightly higher peptidome coverage. Taken together, we propose an optimized informatics pipeline combining MS-GF+ for initial database searching with IQ (or AMT tag) approaches for identification and label-free quantification for high-throughput, comprehensive, and quantitative peptidomics analysis. Graphical Abstract ᅟ.
Keywords: Accurate mass and time tag; Identification; Informed quantitation; MS-GF+; Peptidomics; Quantification.
Figures
References
-
- Schrader M, Schulz-Knappe P. Peptidomics technologies for human body fluids. Trends Biotechnol. 2001;19:S55–60. - PubMed
-
- Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J Neurochem. 2003;87:642–656. - PubMed
-
- Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. - PubMed
-
- Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:451–483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

