Dances with Membranes: Breakthroughs from Super-resolution Imaging
- PMID: 26015281
- PMCID: PMC5584789
- DOI: 10.1016/bs.ctm.2015.03.008
Dances with Membranes: Breakthroughs from Super-resolution Imaging
Abstract
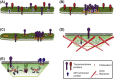
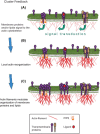
Biological membrane organization mediates numerous cellular functions and has also been connected with an immense number of human diseases. However, until recently, experimental methodologies have been unable to directly visualize the nanoscale details of biological membranes, particularly in intact living cells. Numerous models explaining membrane organization have been proposed, but testing those models has required indirect methods; the desire to directly image proteins and lipids in living cell membranes is a strong motivation for the advancement of technology. The development of super-resolution microscopy has provided powerful tools for quantification of membrane organization at the level of individual proteins and lipids, and many of these tools are compatible with living cells. Previously inaccessible questions are now being addressed, and the field of membrane biology is developing rapidly. This chapter discusses how the development of super-resolution microscopy has led to fundamental advances in the field of biological membrane organization. We summarize the history and some models explaining how proteins are organized in cell membranes, and give an overview of various super-resolution techniques and methods of quantifying super-resolution data. We discuss the application of super-resolution techniques to membrane biology in general, and also with specific reference to the fields of actin and actin-binding proteins, virus infection, mitochondria, immune cell biology, and phosphoinositide signaling. Finally, we present our hopes and expectations for the future of super-resolution microscopy in the field of membrane biology.
Keywords: Actin; Cluster feedback; Domain; FPALM; Lipid; Live cell; PALM; Raft; Review; STORM.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
PALM and STORM: unlocking live-cell super-resolution.Biopolymers. 2011 May;95(5):322-31. doi: 10.1002/bip.21586. Epub 2011 Jan 19. Biopolymers. 2011. PMID: 21254001 Review.
-
Recent advances in super-resolution fluorescence imaging and its applications in biology.J Genet Genomics. 2013 Dec 20;40(12):583-95. doi: 10.1016/j.jgg.2013.11.003. Epub 2013 Nov 23. J Genet Genomics. 2013. PMID: 24377865 Review.
-
Multilayer three-dimensional super resolution imaging of thick biological samples.Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20221-6. doi: 10.1073/pnas.0810636105. Epub 2008 Dec 16. Proc Natl Acad Sci U S A. 2008. PMID: 19088193 Free PMC article.
-
Light Microscopy of Mitochondria at the Nanoscale.Annu Rev Biophys. 2020 May 6;49:289-308. doi: 10.1146/annurev-biophys-121219-081550. Epub 2020 Feb 24. Annu Rev Biophys. 2020. PMID: 32092283 Free PMC article. Review.
-
Super-Resolution Microscopy: Shedding Light on the Cellular Plasma Membrane.Chem Rev. 2017 Jun 14;117(11):7457-7477. doi: 10.1021/acs.chemrev.6b00716. Epub 2017 Feb 17. Chem Rev. 2017. PMID: 28211677 Free PMC article. Review.
Cited by
-
Calcium Sensors as Key Hubs in Plant Responses to Biotic and Abiotic Stresses.Front Plant Sci. 2016 Mar 16;7:327. doi: 10.3389/fpls.2016.00327. eCollection 2016. Front Plant Sci. 2016. PMID: 27014336 Free PMC article. Review.
-
Small-Molecule Modulation of Lipid-Dependent Cellular Processes against Cancer: Fats on the Gunpoint.Biomed Res Int. 2018 Aug 15;2018:6437371. doi: 10.1155/2018/6437371. eCollection 2018. Biomed Res Int. 2018. PMID: 30186863 Free PMC article. Review.
-
Functional link between plasma membrane spatiotemporal dynamics, cancer biology, and dietary membrane-altering agents.Cancer Metastasis Rev. 2018 Sep;37(2-3):519-544. doi: 10.1007/s10555-018-9733-1. Cancer Metastasis Rev. 2018. PMID: 29860560 Free PMC article. Review.
-
Triarylmethane Fluorophores Resistant to Oxidative Photobluing.J Am Chem Soc. 2019 Jan 16;141(2):981-989. doi: 10.1021/jacs.8b11036. Epub 2019 Jan 2. J Am Chem Soc. 2019. PMID: 30562459 Free PMC article.
-
Quantitative FRET Microscopy Reveals a Crucial Role of Cytoskeleton in Promoting PI(4,5)P2 Confinement.Int J Mol Sci. 2021 Oct 29;22(21):11727. doi: 10.3390/ijms222111727. Int J Mol Sci. 2021. PMID: 34769158 Free PMC article.
References
-
- Alberts B. 4th ed. Garland Science; New York: 2002. Molecular biology of the cell.
-
- Almeida P.F.F., Pokorny A., Hinderliter A. Thermodynamics of membrane domains. Biochimica et Biophysica Acta–Biomembranes. 2005;1720(1–2):1–13. (Review) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

