Molecular insight into thiopurine resistance: transcriptomic signature in lymphoblastoid cell lines
- PMID: 26015807
- PMCID: PMC4443628
- DOI: 10.1186/s13073-015-0150-6
Molecular insight into thiopurine resistance: transcriptomic signature in lymphoblastoid cell lines
Abstract
Background: There has been considerable progress in the management of acute lymphoblastic leukemia (ALL) but further improvement is needed to increase long-term survival. The thiopurine agent 6-mercaptopurine (6-MP) used for ALL maintenance therapy has a key influence on clinical outcomes and relapse prevention. Genetic inheritance in thiopurine metabolism plays a major role in interindividual clinical response variability to thiopurines; however, most cases of thiopurine resistance remain unexplained.
Methods: We used lymphoblastoid cell lines (LCLs) from healthy donors, selected for their extreme thiopurine susceptibility. Thiopurine metabolism was characterized by the determination of TPMT and HPRT activity. We performed genome-wide expression profiling in resistant and sensitive cell lines with the goal of elucidating the mechanisms of thiopurine resistance.
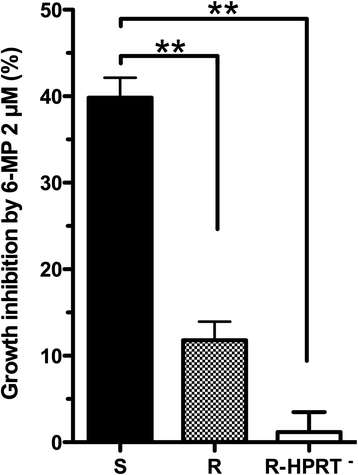
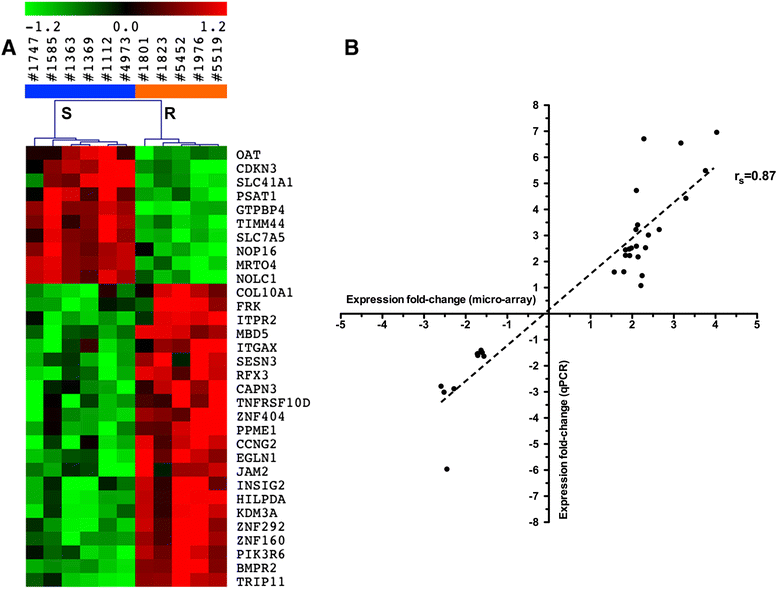
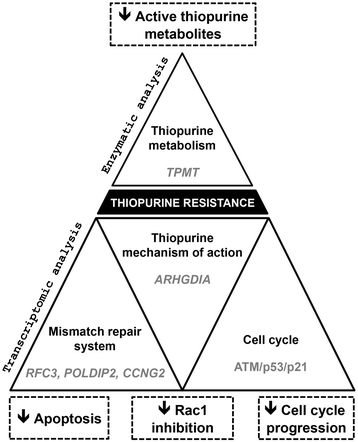
Results: We determined a higher TPMT activity (+44%; P = 0.024) in resistant compared to sensitive cell lines, although there was no difference in HPRT activity. We identified a 32-gene transcriptomic signature that predicts thiopurine resistance. This signature includes the GTPBP4 gene coding for a GTP-binding protein that interacts with p53. A comprehensive pathway analysis of the genes differentially expressed between resistant and sensitive cell lines indicated a role for cell cycle and DNA mismatch repair system in thiopurine resistance. It also revealed overexpression of the ATM/p53/p21 pathway, which is activated in response to DNA damage and induces cell cycle arrest in thiopurine resistant LCLs. Furthermore, overexpression of the p53 target gene TNFRSF10D or the negative cell cycle regulator CCNG2 induces cell cycle arrest and may also contribute to thiopurine resistance. ARHGDIA under-expression in resistant cell lines may constitute a novel molecular mechanism contributing to thiopurine resistance based on Rac1 inhibition induced apoptosis and in relation with thiopurine pharmacodynamics.
Conclusion: Our study provides new insights into the molecular mechanisms underlying thiopurine resistance and suggests a potential research focus for developing tailored medicine.
Figures

Similar articles
-
Thiopurine S-methyltransferase phenotype-genotype correlation in children with acute lymphoblastic leukemia.Acta Pol Pharm. 2012 May-Jun;69(3):405-10. Acta Pol Pharm. 2012. PMID: 22594254
-
Analysis of thiopurine S-methyltransferase polymorphism in the population of Serbia and Montenegro and mercaptopurine therapy tolerance in childhood acute lymphoblastic leukemia.Ther Drug Monit. 2006 Dec;28(6):800-6. doi: 10.1097/01.ftd.0000249947.17676.92. Ther Drug Monit. 2006. PMID: 17164697
-
S-adenosylmethionine regulates thiopurine methyltransferase activity and decreases 6-mercaptopurine cytotoxicity in MOLT lymphoblasts.Biochem Pharmacol. 2009 Jun 15;77(12):1845-53. doi: 10.1016/j.bcp.2009.03.006. Epub 2009 Mar 19. Biochem Pharmacol. 2009. PMID: 19428339
-
The clinical impact of thiopurine methyltransferase polymorphisms on thiopurine treatment.Nucleosides Nucleotides Nucleic Acids. 2004 Oct;23(8-9):1385-91. doi: 10.1081/NCN-200027637. Nucleosides Nucleotides Nucleic Acids. 2004. PMID: 15571264 Review.
-
Review article: the benefits of pharmacogenetics for improving thiopurine therapy in inflammatory bowel disease.Aliment Pharmacol Ther. 2012 Jan;35(1):15-36. doi: 10.1111/j.1365-2036.2011.04905.x. Epub 2011 Nov 2. Aliment Pharmacol Ther. 2012. PMID: 22050052 Review.
Cited by
-
Copy number alterations in B-cell development genes, drug resistance, and clinical outcome in pediatric B-cell precursor acute lymphoblastic leukemia.Sci Rep. 2019 Mar 15;9(1):4634. doi: 10.1038/s41598-019-41078-4. Sci Rep. 2019. PMID: 30874617 Free PMC article.
-
Efforts in redesigning the antileukemic drug 6-thiopurine: decreasing toxic side effects while maintaining efficacy.Medchemcomm. 2018 Dec 20;10(1):169-179. doi: 10.1039/c8md00463c. eCollection 2019 Jan 1. Medchemcomm. 2018. PMID: 30774864 Free PMC article.
-
NOG1 downregulates type I interferon production by targeting phosphorylated interferon regulatory factor 3.PLoS Pathog. 2023 Jul 6;19(7):e1011511. doi: 10.1371/journal.ppat.1011511. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37410776 Free PMC article.
-
Thiopurine methyltransferase activity in children with acute myeloid leukemia.Oncol Lett. 2018 Oct;16(4):4699-4706. doi: 10.3892/ol.2018.9191. Epub 2018 Jul 23. Oncol Lett. 2018. PMID: 30214603 Free PMC article.
-
Metabolism, Biochemical Actions, and Chemical Synthesis of Anticancer Nucleosides, Nucleotides, and Base Analogs.Chem Rev. 2016 Dec 14;116(23):14379-14455. doi: 10.1021/acs.chemrev.6b00209. Epub 2016 Nov 23. Chem Rev. 2016. PMID: 27960273 Free PMC article. Review.
References
-
- Ko RH, Ji L, Barnette P, Bostrom B, Hutchinson R, Raetz E, et al. Outcome of patients treated for relapsed or refractory acute lymphoblastic leukemia: a Therapeutic Advances in Childhood Leukemia Consortium study. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:648–54. doi: 10.1200/JCO.2009.22.2950. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

