SeqGL Identifies Context-Dependent Binding Signals in Genome-Wide Regulatory Element Maps
- PMID: 26016777
- PMCID: PMC4446265
- DOI: 10.1371/journal.pcbi.1004271
SeqGL Identifies Context-Dependent Binding Signals in Genome-Wide Regulatory Element Maps
Abstract
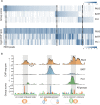
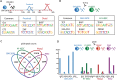
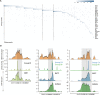
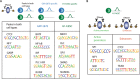
Genome-wide maps of transcription factor (TF) occupancy and regions of open chromatin implicitly contain DNA sequence signals for multiple factors. We present SeqGL, a novel de novo motif discovery algorithm to identify multiple TF sequence signals from ChIP-, DNase-, and ATAC-seq profiles. SeqGL trains a discriminative model using a k-mer feature representation together with group lasso regularization to extract a collection of sequence signals that distinguish peak sequences from flanking regions. Benchmarked on over 100 ChIP-seq experiments, SeqGL outperformed traditional motif discovery tools in discriminative accuracy. Furthermore, SeqGL can be naturally used with multitask learning to identify genomic and cell-type context determinants of TF binding. SeqGL successfully scales to the large multiplicity of sequence signals in DNase- or ATAC-seq maps. In particular, SeqGL was able to identify a number of ChIP-seq validated sequence signals that were not found by traditional motif discovery algorithms. Thus compared to widely used motif discovery algorithms, SeqGL demonstrates both greater discriminative accuracy and higher sensitivity for detecting the DNA sequence signals underlying regulatory element maps. SeqGL is available at http://cbio.mskcc.org/public/Leslie/SeqGL/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Sequence and chromatin determinants of cell-type-specific transcription factor binding.Genome Res. 2012 Sep;22(9):1723-34. doi: 10.1101/gr.127712.111. Genome Res. 2012. PMID: 22955984 Free PMC article.
-
Cell-type specificity of ChIP-predicted transcription factor binding sites.BMC Genomics. 2012 Aug 3;13:372. doi: 10.1186/1471-2164-13-372. BMC Genomics. 2012. PMID: 22863112 Free PMC article.
-
GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding.Bioinformatics. 2016 Feb 15;32(4):490-6. doi: 10.1093/bioinformatics/btv565. Epub 2015 Oct 17. Bioinformatics. 2016. PMID: 26476779 Free PMC article.
-
An algorithmic perspective of de novo cis-regulatory motif finding based on ChIP-seq data.Brief Bioinform. 2018 Sep 28;19(5):1069-1081. doi: 10.1093/bib/bbx026. Brief Bioinform. 2018. PMID: 28334268 Review.
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
Cited by
-
High-Resolution Mapping of Multiway Enhancer-Promoter Interactions Regulating Pathogen Detection.Mol Cell. 2020 Oct 15;80(2):359-373.e8. doi: 10.1016/j.molcel.2020.09.005. Epub 2020 Sep 28. Mol Cell. 2020. PMID: 32991830 Free PMC article.
-
Prediction of regulatory motifs from human Chip-sequencing data using a deep learning framework.Nucleic Acids Res. 2019 Sep 5;47(15):7809-7824. doi: 10.1093/nar/gkz672. Nucleic Acids Res. 2019. PMID: 31372637 Free PMC article.
-
Condition-Specific Modeling of Biophysical Parameters Advances Inference of Regulatory Networks.Cell Rep. 2018 Apr 10;23(2):376-388. doi: 10.1016/j.celrep.2018.03.048. Cell Rep. 2018. PMID: 29641998 Free PMC article.
-
A machine learning-based framework for modeling transcription elongation.Proc Natl Acad Sci U S A. 2021 Feb 9;118(6):e2007450118. doi: 10.1073/pnas.2007450118. Proc Natl Acad Sci U S A. 2021. PMID: 33526657 Free PMC article.
-
Combining signal and sequence to detect RNA polymerase initiation in ATAC-seq data.PLoS One. 2020 Apr 30;15(4):e0232332. doi: 10.1371/journal.pone.0232332. eCollection 2020. PLoS One. 2020. PMID: 32353042 Free PMC article.
References
-
- Brenner, C. HOMER: Software for motif discovery and next-gen sequencing analysis. 2012.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

