Mitochondrial transcript maturation and its disorders
- PMID: 26016801
- PMCID: PMC4493943
- DOI: 10.1007/s10545-015-9859-z
Mitochondrial transcript maturation and its disorders
Abstract
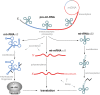
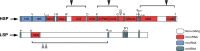
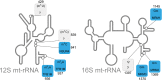
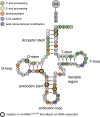
Mitochondrial respiratory chain deficiencies exhibit a wide spectrum of clinical presentations owing to defective mitochondrial energy production through oxidative phosphorylation. These defects can be caused by either mutations in the mitochondrial DNA (mtDNA) or mutations in nuclear genes coding for mitochondrially-targeted proteins. The underlying pathomechanisms can affect numerous pathways involved in mitochondrial biology including expression of mtDNA-encoded genes. Expression of the mitochondrial genes is extensively regulated at the post-transcriptional stage and entails nucleolytic cleavage of precursor RNAs, RNA nucleotide modifications, RNA polyadenylation, RNA quality and stability control. These processes ensure proper mitochondrial RNA (mtRNA) function, and are regulated by dedicated, nuclear-encoded enzymes. Recent growing evidence suggests that mutations in these nuclear genes, leading to incorrect maturation of RNAs, are a cause of human mitochondrial disease. Additionally, mutations in mtDNA-encoded genes may also affect RNA maturation and are frequently associated with human disease. We review the current knowledge on a subset of nuclear-encoded genes coding for proteins involved in mitochondrial RNA maturation, for which genetic variants impacting upon mitochondrial pathophysiology have been reported. Also, primary pathological mtDNA mutations with recognised effects upon RNA processing are described.
Figures


References
-
- Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Reports. 2015;10:920–932. - PubMed
-
- Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013;17:386–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

