A Multiantigenic DNA Vaccine That Induces Broad Hepatitis C Virus-Specific T-Cell Responses in Mice
- PMID: 26018154
- PMCID: PMC4505674
- DOI: 10.1128/JVI.00803-15
A Multiantigenic DNA Vaccine That Induces Broad Hepatitis C Virus-Specific T-Cell Responses in Mice
Abstract
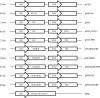
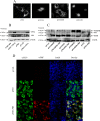
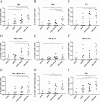
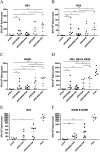
There are 3 to 4 million new hepatitis C virus (HCV) infections annually around the world, but no vaccine is available. Robust T-cell mediated responses are necessary for effective clearance of the virus, and DNA vaccines result in a cell-mediated bias. Adjuvants are often required for effective vaccination, but during natural lytic viral infections damage-associated molecular patterns (DAMPs) are released, which act as natural adjuvants. Hence, a vaccine that induces cell necrosis and releases DAMPs will result in cell-mediated immunity (CMI), similar to that resulting from natural lytic viral infection. We have generated a DNA vaccine with the ability to elicit strong CMI against the HCV nonstructural (NS) proteins (3, 4A, 4B, and 5B) by encoding a cytolytic protein, perforin (PRF), and the antigens on a single plasmid. We examined the efficacy of the vaccines in C57BL/6 mice, as determined by gamma interferon enzyme-linked immunosorbent spot assay, cell proliferation studies, and intracellular cytokine production. Initially, we showed that encoding the NS4A protein in a vaccine which encoded only NS3 reduced the immunogenicity of NS3, whereas including PRF increased NS3 immunogenicity. In contrast, the inclusion of NS4A increased the immunogenicity of the NS3, NS4B, andNS5B proteins, when encoded in a DNA vaccine that also encoded PRF. Finally, vaccines that also encoded PRF elicited similar levels of CMI against each protein after vaccination with DNA encoding NS3, NS4A, NS4B, and NS5B compared to mice vaccinated with DNA encoding only NS3 or NS4B/5B. Thus, we have developed a promising "multiantigen" vaccine that elicits robust CMI.
Importance: Since their development, vaccines have reduced the global burden of disease. One strategy for vaccine development is to use commercially viable DNA technology, which has the potential to generate robust immune responses. Hepatitis C virus causes chronic liver infection and is a leading cause of liver cancer. To date, no vaccine is currently available, and treatment is costly and often results in side effects, limiting the number of patients who are treated. Despite recent advances in treatment, prevention remains the key to efficient control and elimination of this virus. Here, we describe a novel DNA vaccine against hepatitis C virus that is capable of inducing robust cell-mediated immune responses in mice and is a promising vaccine candidate for humans.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Induction of Genotype Cross-Reactive, Hepatitis C Virus-Specific, Cell-Mediated Immunity in DNA-Vaccinated Mice.J Virol. 2018 Mar 28;92(8):e02133-17. doi: 10.1128/JVI.02133-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29437963 Free PMC article.
-
Priming with two DNA vaccines expressing hepatitis C virus NS3 protein targeting dendritic cells elicits superior heterologous protective potential in mice.Arch Virol. 2015 Oct;160(10):2517-24. doi: 10.1007/s00705-015-2535-7. Epub 2015 Jul 28. Arch Virol. 2015. PMID: 26215441
-
Strong HCV NS3/4a, NS4b, NS5a, NS5b-specific cellular immune responses induced in Rhesus macaques by a novel HCV genotype 1a/1b consensus DNA vaccine.Hum Vaccin Immunother. 2014;10(8):2357-65. doi: 10.4161/hv.29590. Hum Vaccin Immunother. 2014. PMID: 25424943 Free PMC article.
-
DNA vaccines.Intervirology. 1999;42(2-3):117-24. doi: 10.1159/000024971. Intervirology. 1999. PMID: 10516466 Review.
-
DNA vaccines for hepatitis C virus.Intervirology. 2001;44(2-3):143-53. doi: 10.1159/000050041. Intervirology. 2001. PMID: 11509875 Review.
Cited by
-
Enhanced T Cell Responses Induced by a Necrotic Dendritic Cell Vaccine, Expressing HCV NS3.Front Microbiol. 2020 Nov 24;11:559105. doi: 10.3389/fmicb.2020.559105. eCollection 2020. Front Microbiol. 2020. PMID: 33343515 Free PMC article.
-
Hepatitis C virus DNA vaccines: a systematic review.Virol J. 2021 Dec 13;18(1):248. doi: 10.1186/s12985-021-01716-8. Virol J. 2021. PMID: 34903252 Free PMC article.
-
Reciprocal Inhibition of Immunogenic Performance in Mice of Two Potent DNA Immunogens Targeting HCV-Related Liver Cancer.Microorganisms. 2021 May 17;9(5):1073. doi: 10.3390/microorganisms9051073. Microorganisms. 2021. PMID: 34067686 Free PMC article.
-
Intradermal delivery of DNA encoding HCV NS3 and perforin elicits robust cell-mediated immunity in mice and pigs.Gene Ther. 2016 Jan;23(1):26-37. doi: 10.1038/gt.2015.86. Epub 2015 Oct 1. Gene Ther. 2016. PMID: 26262584
-
Toward DNA-Based T-Cell Mediated Vaccines to Target HIV-1 and Hepatitis C Virus: Approaches to Elicit Localized Immunity for Protection.Front Cell Infect Microbiol. 2019 Apr 3;9:91. doi: 10.3389/fcimb.2019.00091. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31001491 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

