Discovery of Dengue Virus NS4B Inhibitors
- PMID: 26018165
- PMCID: PMC4524232
- DOI: 10.1128/JVI.00855-15
Discovery of Dengue Virus NS4B Inhibitors
Abstract
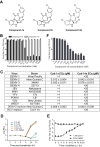
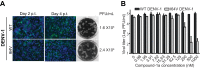
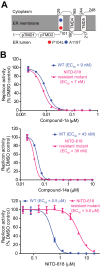
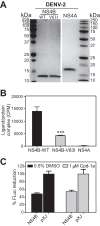
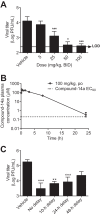
The four serotypes of dengue virus (DENV-1 to -4) represent the most prevalent mosquito-borne viral pathogens in humans. No clinically approved vaccine or antiviral is currently available for DENV. Here we report a spiropyrazolopyridone compound that potently inhibits DENV both in vitro and in vivo. The inhibitor was identified through screening of a 1.8-million-compound library by using a DENV-2 replicon assay. The compound selectively inhibits DENV-2 and -3 (50% effective concentration [EC50], 10 to 80 nM) but not DENV-1 and -4 (EC50,>20 M). Resistance analysis showed that a mutation at amino acid 63 of DENV-2 NS4B (a nonenzymatic transmembrane protein and a component of the viral replication complex) could confer resistance to compound inhibition. Genetic studies demonstrate that variations at amino acid 63 of viral NS4B are responsible for the selective inhibition of DENV-2 and -3. Medicinal chemistry improved the physicochemical properties of the initial “hit” (compound 1), leading to compound 14a, which has good in vivo pharmacokinetics. Treatment of DENV-2-infected AG129 mice with compound 14a suppressed viremia, even when the treatment started after viral infection. The results have proven the concept that inhibitors of NS4B could potentially be developed for clinical treatment of DENV infection. Compound 14a represents a potential preclinical candidate for treatment of DENV-2- and -3-infected patients.
Figures


References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi:10.1038/nature12060. - DOI - PMC - PubMed
-
- Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. doi:10.1016/S0140-6736(12)61428-7. - DOI - PubMed
-
- Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon IK, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A, CYD14 Study Group. 2014. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384:1358–1365. doi:10.1016/S0140-6736(14)61060-6. - DOI - PubMed
-
- Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramirez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortes Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F, CYD15 Study Group. 2015. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med 372:113–123. doi:10.1056/NEJMoa1411037. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

