Functional Analysis of the 60-Nucleotide Duplication in the Respiratory Syncytial Virus Buenos Aires Strain Attachment Glycoprotein
- PMID: 26018171
- PMCID: PMC4524256
- DOI: 10.1128/JVI.01045-15
Functional Analysis of the 60-Nucleotide Duplication in the Respiratory Syncytial Virus Buenos Aires Strain Attachment Glycoprotein
Abstract
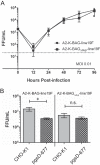
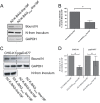
There are two subgroups of respiratory syncytial virus (RSV), A and B, and within each subgroup, isolates are further divided into clades. Several years ago, multiple subgroup B isolates which contained a duplication of 60 nucleotides in the glycoprotein (G) gene were described. These isolates were given a new clade designation of BA based on the site of isolation, Buenos Aires, Argentina. BA RSV strains have since become the predominant circulating clade of RSV B viruses. We hypothesized that the duplicated region in G serves to enhance the function of G in the virus life cycle. We generated recombinant viruses that express a consensus BA G gene or a consensus BA G gene lacking the duplication (GΔdup). We determined that the duplicated region functions during virus attachment to cells. Additionally, we showed that in vitro, the virus containing the duplication has a fitness advantage compared to the virus without the duplication. Our data demonstrate that the duplicated region in the BA strain G protein augments virus attachment and fitness.
Importance: Respiratory syncytial virus (RSV) is an important pathogen for infants for which there is no vaccine. Different strains of RSV circulate from year to year, and the predominating strains change over time. Subgroup B RSV strains with a duplication in the attachment glycoprotein (G) emerged and then became the dominant B genotype. We found that a recombinant virus harboring the duplication bound more efficiently to cells and was more fit than a recombinant strain lacking the duplication. Our work advances a mechanism for an important natural RSV mutation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Collins PL, Crowe JE Jr. 2007. Respiratory syncytial virus and metapneumovirus, p 1601–1646. In Knipe DM, Howley PM (ed), Fields virology, vol 2 Lippincott, Williams, and Wilkins, Philadelphia, PA.
-
- Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 79(Pt 9):2221–2229. - PubMed
-
- Trento A, Viegas M, Galiano M, Videla C, Carballal G, Mistchenko AS, Melero JA. 2006. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol 80:975–984. doi:10.1128/JVI.80.2.975-984.2006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

