Whole transcriptome analysis with sequencing: methods, challenges and potential solutions
- PMID: 26018601
- PMCID: PMC6233721
- DOI: 10.1007/s00018-015-1934-y
Whole transcriptome analysis with sequencing: methods, challenges and potential solutions
Abstract
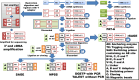
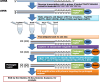
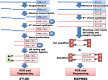
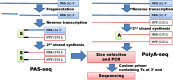
Whole transcriptome analysis plays an essential role in deciphering genome structure and function, identifying genetic networks underlying cellular, physiological, biochemical and biological systems and establishing molecular biomarkers that respond to diseases, pathogens and environmental challenges. Here, we review transcriptome analysis methods and technologies that have been used to conduct whole transcriptome shotgun sequencing or whole transcriptome tag/target sequencing analyses. We focus on how adaptors/linkers are added to both 5' and 3' ends of mRNA molecules for cloning or PCR amplification before sequencing. Challenges and potential solutions are also discussed. In brief, next generation sequencing platforms have accelerated releases of the large amounts of gene expression data. It is now time for the genome research community to assemble whole transcriptomes of all species and collect signature targets for each gene/transcript, and thus use known genes/transcripts to determine known transcriptomes directly in the near future.
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures


References
-
- Granjeaud S, Bertucci F, Jordan BR. Expression profiling: DNA arrays in many guises. Bioessays. 1999;21(9):781–790. - PubMed
-
- Altman RB, Raychaudhuri S. Whole-genome expression analysis: challenges beyond clustering. Curr Opin Struct Biol. 2001;11(3):340–347. - PubMed
-
- Hsiao LL, Stears RL, Hong RL, Gullans SR. Prospective use of DNA microarrays for evaluating renal function and disease. Curr Opin Nephrol Hypertens. 2000;9(3):253–258. - PubMed
-
- Celis JE, Kruhøffer M, Gromova I, Frederiksen C, Ostergaard M, Thykjaer T, Gromov P, Yu J, Pálsdóttir H, Magnusson N, Orntoft TF. Gene expression profiling: monitoring transcription and translation products using DNA microarrays and proteomics. FEBS Lett. 2000;480(1):2–16. - PubMed
-
- Manger ID, Relman DA. How the host ‘sees’ pathogens: global gene expression responses to infection. Curr Opin Immunol. 2000;12(2):215–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

