JAFFA: High sensitivity transcriptome-focused fusion gene detection
- PMID: 26019724
- PMCID: PMC4445815
- DOI: 10.1186/s13073-015-0167-x
JAFFA: High sensitivity transcriptome-focused fusion gene detection
Abstract
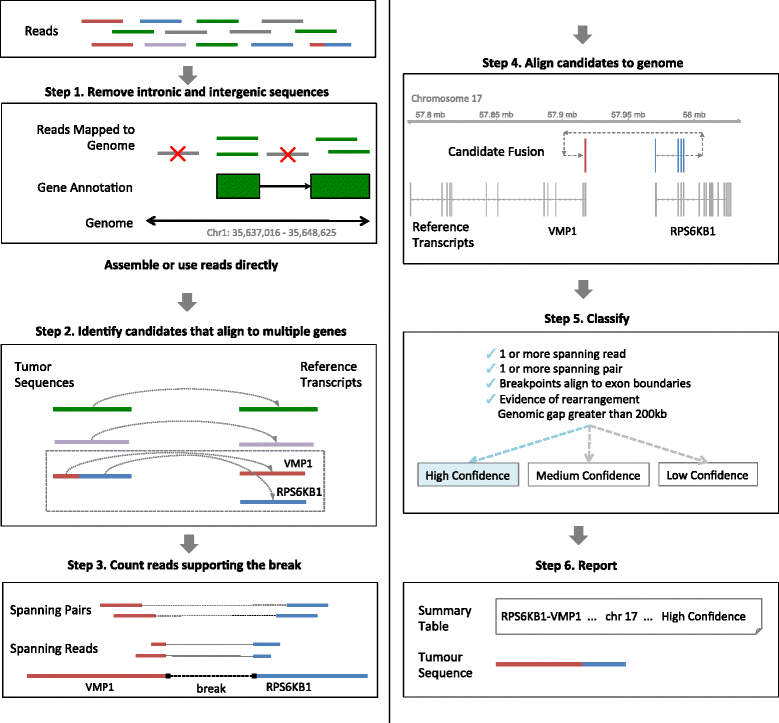
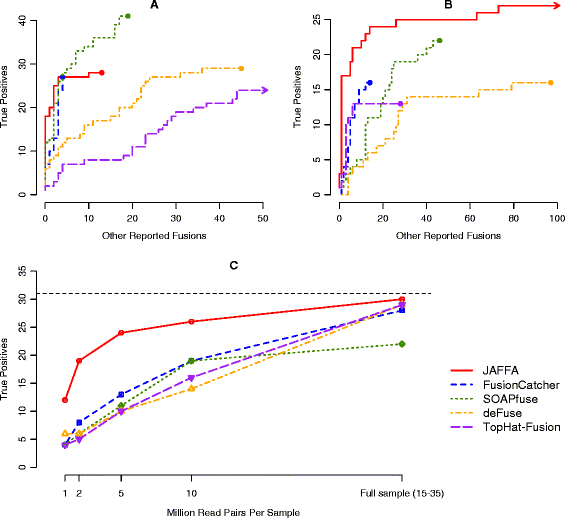
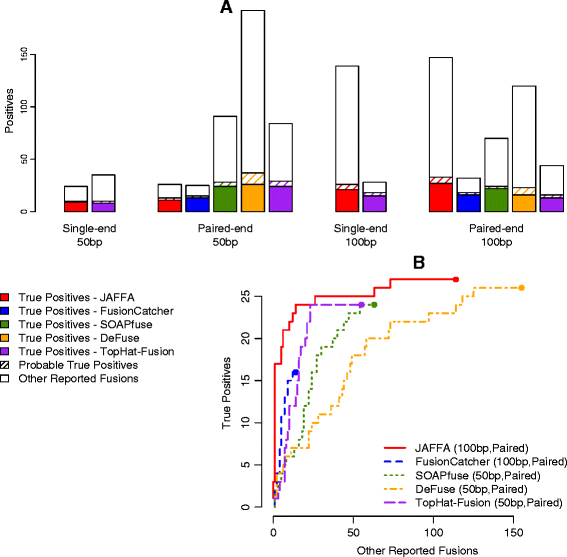
Genomic instability is a hallmark of cancer and, as such, structural alterations and fusion genes are common events in the cancer landscape. RNA sequencing (RNA-Seq) is a powerful method for profiling cancers, but current methods for identifying fusion genes are optimised for short reads. JAFFA (https://github.com/Oshlack/JAFFA/wiki) is a sensitive fusion detection method that outperforms other methods with reads of 100 bp or greater. JAFFA compares a cancer transcriptome to the reference transcriptome, rather than the genome, where the cancer transcriptome is inferred using long reads directly or by de novo assembling short reads.
Figures
References
-
- Edwards PAW. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220:244–54. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

