Characterization of the Escherichia coli σ(S) core regulon by Chromatin Immunoprecipitation-sequencing (ChIP-seq) analysis
- PMID: 26020590
- PMCID: PMC4447067
- DOI: 10.1038/srep10469
Characterization of the Escherichia coli σ(S) core regulon by Chromatin Immunoprecipitation-sequencing (ChIP-seq) analysis
Abstract
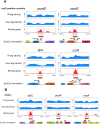
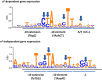
In bacteria, selective promoter recognition by RNA polymerase is achieved by its association with σ factors, accessory subunits able to direct RNA polymerase "core enzyme" (E) to different promoter sequences. Using Chromatin Immunoprecipitation-sequencing (ChIP-seq), we searched for promoters bound by the σ(S)-associated RNA polymerase form (Eσ(S)) during transition from exponential to stationary phase. We identified 63 binding sites for Eσ(S) overlapping known or putative promoters, often located upstream of genes (encoding either ORFs or non-coding RNAs) showing at least some degree of dependence on the σ(S)-encoding rpoS gene. Eσ(S) binding did not always correlate with an increase in transcription level, suggesting that, at some σ(S)-dependent promoters, Eσ(S) might remain poised in a pre-initiation state upon binding. A large fraction of Eσ(S)-binding sites corresponded to promoters recognized by RNA polymerase associated with σ(70) or other σ factors, suggesting a considerable overlap in promoter recognition between different forms of RNA polymerase. In particular, Eσ(S) appears to contribute significantly to transcription of genes encoding proteins involved in LPS biosynthesis and in cell surface composition. Finally, our results highlight a direct role of Eσ(S) in the regulation of non coding RNAs, such as OmrA/B, RyeA/B and SibC.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Lloyd G., Landini P. & Busby S. Activation and repression of transcription initiation in bacteria. Essays Biochem 37, 17–31 (2001). - PubMed
-
- Hengge-Aronis R. in Bacterial Stress Responses 161–178 (ASM Press, Washington, DC; 2000).
-
- Landini P., Egli T., Wolf J. & Lacour S. sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ Microbiol Rep 6, 1–13 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

