Transcriptome outlier analysis implicates schizophrenia susceptibility genes and enriches putatively functional rare genetic variants
- PMID: 26022996
- PMCID: PMC4512633
- DOI: 10.1093/hmg/ddv199
Transcriptome outlier analysis implicates schizophrenia susceptibility genes and enriches putatively functional rare genetic variants
Abstract
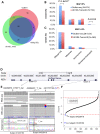
We searched a gene expression dataset comprised of 634 schizophrenia (SZ) cases and 713 controls for expression outliers (i.e., extreme tails of the distribution of transcript expression values) with SZ cases overrepresented compared with controls. These outlier genes were enriched for brain expression and for genes known to be associated with neurodevelopmental disorders. SZ cases showed higher outlier burden (i.e., total outlier events per subject) than controls for genes within copy number variants (CNVs) associated with SZ or neurodevelopmental disorders. Outlier genes were enriched for CNVs and for rare putative regulatory variants, but this only explained a small proportion of the outlier subjects, highlighting the underlying presence of additional genetic and potentially, epigenetic mechanisms.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- McGuffin P., Farmer A.E., Gottesman I.I., Murray R.M., Reveley A.M. (1984) Twin concordance for operationally defined schizophrenia. Confirmation of familiality and heritability. Arch. Gen. Psychiatry, 41, 541–545. - PubMed
Publication types
MeSH terms
Grants and funding
- R01MH060870/MH/NIMH NIH HHS/United States
- U01MH079469/MH/NIMH NIH HHS/United States
- U01 MH046276/MH/NIMH NIH HHS/United States
- R01MH059588/MH/NIMH NIH HHS/United States
- U01MH079470/MH/NIMH NIH HHS/United States
- R01 MH067257/MH/NIMH NIH HHS/United States
- R01MH059566/MH/NIMH NIH HHS/United States
- U01 MH079470/MH/NIMH NIH HHS/United States
- R01 MH059586/MH/NIMH NIH HHS/United States
- WT091310/WT_/Wellcome Trust/United Kingdom
- R01MH059586/MH/NIMH NIH HHS/United States
- R01 MH061675/MH/NIMH NIH HHS/United States
- R01MH094091/MH/NIMH NIH HHS/United States
- R21 MH102685/MH/NIMH NIH HHS/United States
- R01MH059587/MH/NIMH NIH HHS/United States
- U01 MH079469/MH/NIMH NIH HHS/United States
- R01 MH094116/MH/NIMH NIH HHS/United States
- R01 MH060870/MH/NIMH NIH HHS/United States
- R01 MH081800/MH/NIMH NIH HHS/United States
- R01 MH059571/MH/NIMH NIH HHS/United States
- R01 MH059565/MH/NIMH NIH HHS/United States
- R01MH067257/MH/NIMH NIH HHS/United States
- R01 MH059587/MH/NIMH NIH HHS/United States
- R01 MH094091/MH/NIMH NIH HHS/United States
- U01MH046276/MH/NIMH NIH HHS/United States
- R01MH061675/MH/NIMH NIH HHS/United States
- RC2MH090030/MH/NIMH NIH HHS/United States
- R01MH059571/MH/NIMH NIH HHS/United States
- R01MH060879/MH/NIMH NIH HHS/United States
- R01 MH059566/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01MH094116/MH/NIMH NIH HHS/United States
- R01 MH059588/MH/NIMH NIH HHS/United States
- R01MH081800/MH/NIMH NIH HHS/United States
- RC2 MH090030/MH/NIMH NIH HHS/United States
- R01MH059565/MH/NIMH NIH HHS/United States
- R01 MH060879/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

