3D Imaging of Axons in Transparent Spinal Cords from Rodents and Nonhuman Primates
- PMID: 26023683
- PMCID: PMC4444235
- DOI: 10.1523/ENEURO.0001-15.2015
3D Imaging of Axons in Transparent Spinal Cords from Rodents and Nonhuman Primates
Abstract
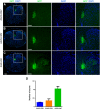
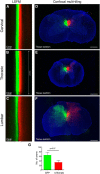
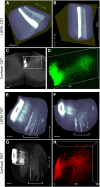
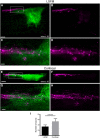
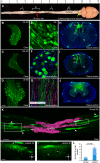
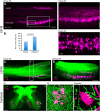
The histological assessment of spinal cord tissue in three dimensions has previously been very time consuming and prone to errors of interpretation. Advances in tissue clearing have significantly improved visualization of fluorescently labelled axons. While recent proof-of-concept studies have been performed with transgenic mice in which axons were prelabeled with GFP, investigating axonal regeneration requires stringent axonal tracing methods as well as the use of animal models in which transgenic axonal labeling is not available. Using rodent models of spinal cord injury, we labeled axon tracts of interest using both adeno-associated virus and chemical tracers and performed tetrahydrofuran-based tissue clearing to image multiple axon types in spinal cords using light sheet and confocal microscopy. Using this approach, we investigated the relationships between axons and scar-forming cells at the injury site as well as connections between sensory axons and motor pools in the spinal cord. In addition, we used these methods to trace axons in nonhuman primates. This reproducible and adaptable virus-based approach can be combined with transgenic mice or with chemical-based tract-tracing methods, providing scientists with flexibility in obtaining axonal trajectory information from transparent tissue.
Keywords: 3DISCO; axon regeneration; spinal cord injury; tissue clearing.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Blits B, Derks S, Twisk J, Ehlert E, Prins J, Verhaagen J (2010) Adeno-associated viral vector (AAV)-mediated gene transfer in the red nucleus of the adult rat brain: comparative analysis of the transduction properties of seven AAV serotypes and lentiviral vectors. J Neurosci Methods 185:257-263. 10.1016/j.jneumeth.2009.10.009 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
