Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
- PMID: 26023780
- PMCID: PMC4449185
- DOI: 10.1371/journal.ppat.1004932
Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
Abstract
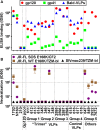
Eliciting broad tier 2 neutralizing antibodies (nAbs) is a major goal of HIV-1 vaccine research. Here we investigated the ability of native, membrane-expressed JR-FL Env trimers to elicit nAbs. Unusually potent nAb titers developed in 2 of 8 rabbits immunized with virus-like particles (VLPs) expressing trimers (trimer VLP sera) and in 1 of 20 rabbits immunized with DNA expressing native Env trimer, followed by a protein boost (DNA trimer sera). All 3 sera neutralized via quaternary epitopes and exploited natural gaps in the glycan defenses of the second conserved region of JR-FL gp120. Specifically, trimer VLP sera took advantage of the unusual absence of a glycan at residue 197 (present in 98.7% of Envs). Intriguingly, removing the N197 glycan (with no loss of tier 2 phenotype) rendered 50% or 16.7% (n = 18) of clade B tier 2 isolates sensitive to the two trimer VLP sera, showing broad neutralization via the surface masked by the N197 glycan. Neutralizing sera targeted epitopes that overlap with the CD4 binding site, consistent with the role of the N197 glycan in a putative "glycan fence" that limits access to this region. A bioinformatics analysis suggested shared features of one of the trimer VLP sera and monoclonal antibody PG9, consistent with its trimer-dependency. The neutralizing DNA trimer serum took advantage of the absence of a glycan at residue 230, also proximal to the CD4 binding site and suggesting an epitope similar to that of monoclonal antibody 8ANC195, albeit lacking tier 2 breadth. Taken together, our data show for the first time that strain-specific holes in the glycan fence can allow the development of tier 2 neutralizing antibodies to native spikes. Moreover, cross-neutralization can occur in the absence of protecting glycan. Overall, our observations provide new insights that may inform the future development of a neutralizing antibody vaccine.
Conflict of interest statement
The authors have declared that no competing interests exist. Some authors are employed with Altravax Inc, but this does not affect competing interests. No employment, consultancies, patents or products in development or on market represent a competing interest with this article. This does not alter our adherence to all PLOS Pathogens policies on sharing data and materials.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

