A Method for 3D Histopathology Reconstruction Supporting Mouse Microvasculature Analysis
- PMID: 26024221
- PMCID: PMC4449209
- DOI: 10.1371/journal.pone.0126817
A Method for 3D Histopathology Reconstruction Supporting Mouse Microvasculature Analysis
Abstract
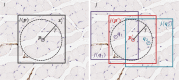
Structural abnormalities of the microvasculature can impair perfusion and function. Conventional histology provides good spatial resolution with which to evaluate the microvascular structure but affords no 3-dimensional information; this limitation could lead to misinterpretations of the complex microvessel network in health and disease. The objective of this study was to develop and evaluate an accurate, fully automated 3D histology reconstruction method to visualize the arterioles and venules within the mouse hind-limb. Sections of the tibialis anterior muscle from C57BL/J6 mice (both normal and subjected to femoral artery excision) were reconstructed using pairwise rigid and affine registrations of 5 µm-thick, paraffin-embedded serial sections digitized at 0.25 µm/pixel. Low-resolution intensity-based rigid registration was used to initialize the nucleus landmark-based registration, and conventional high-resolution intensity-based registration method. The affine nucleus landmark-based registration was developed in this work and was compared to the conventional affine high-resolution intensity-based registration method. Target registration errors were measured between adjacent tissue sections (pairwise error), as well as with respect to a 3D reference reconstruction (accumulated error, to capture propagation of error through the stack of sections). Accumulated error measures were lower (p < 0.01) for the nucleus landmark technique and superior vasculature continuity was observed. These findings indicate that registration based on automatic extraction and correspondence of small, homologous landmarks may support accurate 3D histology reconstruction. This technique avoids the otherwise problematic "banana-into-cylinder" effect observed using conventional methods that optimize the pairwise alignment of salient structures, forcing them to be section-orthogonal. This approach will provide a valuable tool for high-accuracy 3D histology tissue reconstructions for analysis of diseased microvasculature.
Conflict of interest statement
Figures



References
-
- Csernus B, Ficsor L, Molnar B, Sapi Z. Comparison of the vasculature of myxofibrosarcoma and myxoid liposarcoma using 3d histological reconstruction. Histopathology. 2008;53(Journal Article):410–1.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

