TMEM120A and B: Nuclear Envelope Transmembrane Proteins Important for Adipocyte Differentiation
- PMID: 26024229
- PMCID: PMC4449205
- DOI: 10.1371/journal.pone.0127712
TMEM120A and B: Nuclear Envelope Transmembrane Proteins Important for Adipocyte Differentiation
Abstract
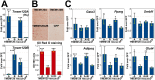
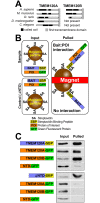
Recent work indicates that the nuclear envelope is a major signaling node for the cell that can influence tissue differentiation processes. Here we present two nuclear envelope trans-membrane proteins TMEM120A and TMEM120B that are paralogs encoded by the Tmem120A and Tmem120B genes. The TMEM120 proteins are expressed preferentially in fat and both are induced during 3T3-L1 adipocyte differentiation. Knockdown of one or the other protein altered expression of several genes required for adipocyte differentiation, Gata3, Fasn, Glut4, while knockdown of both together additionally affected Pparg and Adipoq. The double knockdown also increased the strength of effects, reducing for example Glut4 levels by 95% compared to control 3T3-L1 cells upon pharmacologically induced differentiation. Accordingly, TMEM120A and B knockdown individually and together impacted on adipocyte differentiation/metabolism as measured by lipid accumulation through binding of Oil Red O and coherent anti-Stokes Raman scattering microscopy (CARS). The nuclear envelope is linked to several lipodystrophies through mutations in lamin A; however, lamin A is widely expressed. Thus it is possible that the TMEM120A and B fat-specific nuclear envelope transmembrane proteins may play a contributory role in the tissue-specific pathology of this disorder or in the wider problem of obesity.
Conflict of interest statement
Figures

Similar articles
-
Suppression of adipocyte differentiation and lipid accumulation by stearidonic acid (SDA) in 3T3-L1 cells.Lipids Health Dis. 2017 Sep 25;16(1):181. doi: 10.1186/s12944-017-0574-7. Lipids Health Dis. 2017. PMID: 28946872 Free PMC article.
-
Characterization of a novel murine preadipocyte line, AP-18, isolated from subcutaneous tissue: analysis of adipocyte-related gene expressions.Cell Biol Int. 2010 Feb 22;34(3):293-9. doi: 10.1042/CBI20090063. Cell Biol Int. 2010. PMID: 19947910
-
α, β-Amyrin, a pentacyclic triterpenoid from Protium heptaphyllum suppresses adipocyte differentiation accompanied by down regulation of PPARγ and C/EBPα in 3T3-L1 cells.Biomed Pharmacother. 2019 Jan;109:1860-1866. doi: 10.1016/j.biopha.2018.11.027. Epub 2018 Nov 26. Biomed Pharmacother. 2019. PMID: 30551441
-
The curious case of TMEM120A: Mechanosensor, fat regulator, or antiviral defender?Bioessays. 2022 Jun;44(6):e2200045. doi: 10.1002/bies.202200045. Epub 2022 Apr 14. Bioessays. 2022. PMID: 35419854 Review.
-
TMEM120A/TACAN: A putative regulator of ion channels, mechanosensation, and lipid metabolism.Channels (Austin). 2023 Dec;17(1):2237306. doi: 10.1080/19336950.2023.2237306. Channels (Austin). 2023. PMID: 37523628 Free PMC article. Review.
Cited by
-
Phosphatidic acid is an endogenous negative regulator of PIEZO2 channels and mechanical sensitivity.Nat Commun. 2024 Aug 15;15(1):7020. doi: 10.1038/s41467-024-51181-4. Nat Commun. 2024. PMID: 39147733 Free PMC article.
-
Profiling Cellular Processes in Adipose Tissue during Weight Loss Using Time Series Gene Expression.Genes (Basel). 2018 Oct 29;9(11):525. doi: 10.3390/genes9110525. Genes (Basel). 2018. PMID: 30380678 Free PMC article.
-
TMEM120A/TACAN inhibits mechanically activated PIEZO2 channels.J Gen Physiol. 2022 Aug 1;154(8):e202213164. doi: 10.1085/jgp.202213164. Epub 2022 Jul 12. J Gen Physiol. 2022. PMID: 35819364 Free PMC article.
-
The Cutting Edge: The Role of mTOR Signaling in Laminopathies.Int J Mol Sci. 2019 Feb 15;20(4):847. doi: 10.3390/ijms20040847. Int J Mol Sci. 2019. PMID: 30781376 Free PMC article. Review.
-
Altered adipocyte differentiation and unbalanced autophagy in type 2 Familial Partial Lipodystrophy: an in vitro and in vivo study of adipose tissue browning.Exp Mol Med. 2019 Aug 2;51(8):1-17. doi: 10.1038/s12276-019-0289-0. Exp Mol Med. 2019. PMID: 31375660 Free PMC article.
References
-
- WHO. Obesity and overweight [Internet]. 2011. Available: http://www.who.int.mediacentre/factsheets/fs311/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

