Structural-Functional Analysis Reveals a Specific Domain Organization in Family GH20 Hexosaminidases
- PMID: 26024355
- PMCID: PMC4449183
- DOI: 10.1371/journal.pone.0128075
Structural-Functional Analysis Reveals a Specific Domain Organization in Family GH20 Hexosaminidases
Abstract
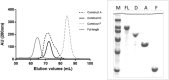
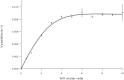
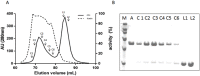
Hexosaminidases are involved in important biological processes catalyzing the hydrolysis of N-acetyl-hexosaminyl residues in glycosaminoglycans and glycoconjugates. The GH20 enzymes present diverse domain organizations for which we propose two minimal model architectures: Model A containing at least a non-catalytic GH20b domain and the catalytic one (GH20) always accompanied with an extra α-helix (GH20b-GH20-α), and Model B with only the catalytic GH20 domain. The large Bifidobacterium bifidum lacto-N-biosidase was used as a model protein to evaluate the minimal functional unit due to its interest and structural complexity. By expressing different truncated forms of this enzyme, we show that Model A architectures cannot be reduced to Model B. In particular, there are two structural requirements general to GH20 enzymes with Model A architecture. First, the non-catalytic domain GH20b at the N-terminus of the catalytic GH20 domain is required for expression and seems to stabilize it. Second, the substrate-binding cavity at the GH20 domain always involves a remote element provided by a long loop from the catalytic domain itself or, when this loop is short, by an element from another domain of the multidomain structure or from the dimeric partner. Particularly, the lacto-N-biosidase requires GH20b and the lectin-like domain at the N- and C-termini of the catalytic GH20 domain to be fully soluble and functional. The lectin domain provides this remote element to the active site. We demonstrate restoration of activity of the inactive GH20b-GH20-α construct (model A architecture) by a complementation assay with the lectin-like domain. The engineering of minimal functional units of multidomain GH20 enzymes must consider these structural requirements.
Conflict of interest statement
Figures




Similar articles
-
Transglycosylation Activity of Engineered Bifidobacterium Lacto-N-Biosidase Mutants at Donor Subsites for Lacto-N-Tetraose Synthesis.Int J Mol Sci. 2021 Mar 22;22(6):3230. doi: 10.3390/ijms22063230. Int J Mol Sci. 2021. PMID: 33810098 Free PMC article.
-
Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum.J Biol Chem. 2013 Apr 26;288(17):11795-806. doi: 10.1074/jbc.M112.420109. Epub 2013 Mar 11. J Biol Chem. 2013. PMID: 23479733 Free PMC article.
-
Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression.J Biol Chem. 2013 Aug 30;288(35):25194-25206. doi: 10.1074/jbc.M113.484733. Epub 2013 Jul 10. J Biol Chem. 2013. PMID: 23843461 Free PMC article.
-
Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins.J Biosci Bioeng. 2005 May;99(5):457-65. doi: 10.1263/jbb.99.457. J Biosci Bioeng. 2005. PMID: 16233817 Review.
-
Structural Insights into the Molecular Evolution of the Archaeal Exo-β-d-Glucosaminidase.Int J Mol Sci. 2019 May 18;20(10):2460. doi: 10.3390/ijms20102460. Int J Mol Sci. 2019. PMID: 31109049 Free PMC article. Review.
Cited by
-
Transglycosylation Activity of Engineered Bifidobacterium Lacto-N-Biosidase Mutants at Donor Subsites for Lacto-N-Tetraose Synthesis.Int J Mol Sci. 2021 Mar 22;22(6):3230. doi: 10.3390/ijms22063230. Int J Mol Sci. 2021. PMID: 33810098 Free PMC article.
-
Proteomic Analysis Revealed the Fruiting-Body Protein Profile of Auricularia polytricha.Curr Microbiol. 2017 Aug;74(8):943-951. doi: 10.1007/s00284-017-1268-0. Epub 2017 May 29. Curr Microbiol. 2017. PMID: 28555376
-
A Second β-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.J Biol Chem. 2015 Dec 25;290(52):30888-900. doi: 10.1074/jbc.M115.688630. Epub 2015 Oct 21. J Biol Chem. 2015. PMID: 26491009 Free PMC article.
-
Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila β-N-acetylhexosaminidase Am2136.Gut Microbes. 2022 Jan-Dec;14(1):2143221. doi: 10.1080/19490976.2022.2143221. Gut Microbes. 2022. PMID: 36394293 Free PMC article.
-
Genomic and transcriptomic landscapes and evolutionary dynamics of molluscan glycoside hydrolase families with implications for algae-feeding biology.Comput Struct Biotechnol J. 2020 Sep 28;18:2744-2756. doi: 10.1016/j.csbj.2020.09.021. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33101612 Free PMC article.
References
-
- Liu T, Yan J, Yang Q. Comparative biochemistry of GH3, GH20 and GH84 beta-N-acetyl-D-hexosaminidases and recent progress in selective inhibitor discovery. Curr Drug Targets. 2012;13: 512–525. - PubMed
-
- Liu T, Chen L, Ma Q, Shen X, Yang Q. Structural Insights into Chitinolytic Enzymes and Inhibition Mechanisms of Selective Inhibitors. Curr Pharm Des. 2013. - PubMed
-
- Rountree JS, Butters TD, Wormald MR, Boomkamp SD, Dwek RA, Asano N et al. Design, synthesis, and biological evaluation of enantiomeric beta-N-acetylhexosaminidase inhibitors LABNAc and DABNAc as potential agents against Tay-Sachs and Sandhoff disease. ChemMedChem. 2009;4: 378–392. 10.1002/cmdc.200800350 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

