The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
- PMID: 26024477
- PMCID: PMC4449002
- DOI: 10.1371/journal.ppat.1004910
The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
Abstract
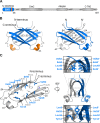
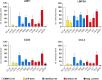
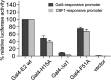
Epstein-Barr virus (EBV) is a γ-herpesvirus that may cause infectious mononucleosis in young adults. In addition, epidemiological and molecular evidence links EBV to the pathogenesis of lymphoid and epithelial malignancies. EBV has the unique ability to transform resting B cells into permanently proliferating, latently infected lymphoblastoid cell lines. Epstein-Barr virus nuclear antigen 2 (EBNA-2) is a key regulator of viral and cellular gene expression for this transformation process. The N-terminal region of EBNA-2 comprising residues 1-58 appears to mediate multiple molecular functions including self-association and transactivation. However, it remains to be determined if the N-terminus of EBNA-2 directly provides these functions or if these activities merely depend on the dimerization involving the N-terminal domain. To address this issue, we determined the three-dimensional structure of the EBNA-2 N-terminal dimerization (END) domain by heteronuclear NMR-spectroscopy. The END domain monomer comprises a small fold of four β-strands and an α-helix which form a parallel dimer by interaction of two β-strands from each protomer. A structure-guided mutational analysis showed that hydrophobic residues in the dimer interface are required for self-association in vitro. Importantly, these interface mutants also displayed severely impaired self-association and transactivation in vivo. Moreover, mutations of solvent-exposed residues or deletion of the α-helix do not impair dimerization but strongly affect the functional activity, suggesting that the EBNA-2 dimer presents a surface that mediates functionally important intra- and/or intermolecular interactions. Our study shows that the END domain is a novel dimerization fold that is essential for functional activity. Since this specific fold is a unique feature of EBNA-2 it might provide a novel target for anti-viral therapeutics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif.J Virol. 1993 Jun;67(6):2990-3003. doi: 10.1128/JVI.67.6.2990-3003.1993. J Virol. 1993. PMID: 8388484 Free PMC article.
-
Biophysical and mutational analysis of the putative bZIP domain of Epstein-Barr virus EBNA 3C.J Virol. 2004 Sep;78(17):9431-45. doi: 10.1128/JVI.78.17.9431-9445.2004. J Virol. 2004. PMID: 15308737 Free PMC article.
-
The conserved domain CR2 of Epstein-Barr virus nuclear antigen leader protein is responsible not only for nuclear matrix association but also for nuclear localization.Virology. 2001 Jan 20;279(2):401-13. doi: 10.1006/viro.2000.0715. Virology. 2001. PMID: 11162796
-
Conserved region CR2 of Epstein-Barr virus nuclear antigen leader protein is a multifunctional domain that mediates self-association as well as nuclear localization and nuclear matrix association.J Virol. 2002 Feb;76(3):1025-32. doi: 10.1128/jvi.76.3.1025-1032.2002. J Virol. 2002. PMID: 11773378 Free PMC article.
-
A single amino acid in EBNA-2 determines superior B lymphoblastoid cell line growth maintenance by Epstein-Barr virus type 1 EBNA-2.J Virol. 2014 Aug;88(16):8743-53. doi: 10.1128/JVI.01000-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850736 Free PMC article.
Cited by
-
EBNA2-EBF1 complexes promote MYC expression and metabolic processes driving S-phase progression of Epstein-Barr virus-infected B cells.Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2200512119. doi: 10.1073/pnas.2200512119. Epub 2022 Jul 20. Proc Natl Acad Sci U S A. 2022. PMID: 35857872 Free PMC article.
-
Epstein-Barr Virus B Cell Growth Transformation: The Nuclear Events.Viruses. 2023 Mar 24;15(4):832. doi: 10.3390/v15040832. Viruses. 2023. PMID: 37112815 Free PMC article. Review.
-
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts EBNA2 activity.PLoS Pathog. 2019 Jul 8;15(7):e1007458. doi: 10.1371/journal.ppat.1007458. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31283782 Free PMC article.
-
EBF1 binds to EBNA2 and promotes the assembly of EBNA2 chromatin complexes in B cells.PLoS Pathog. 2017 Oct 2;13(10):e1006664. doi: 10.1371/journal.ppat.1006664. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28968461 Free PMC article.
-
REST Is Restless in Neuronal and Non-Neuronal Virus Infections: An In Silico Analysis-Based Perspective.Viruses. 2025 Feb 8;17(2):234. doi: 10.3390/v17020234. Viruses. 2025. PMID: 40006989 Free PMC article. Review.
References
-
- Longenecker RM, Kieff E, Cohen JI. Epstein-Barr virus In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, et al., editors. Fields Virology. 2 6 ed. Philadelphia: Lippincott Williams and Wilkins; 2013. p. 1898–959.
-
- Adldinger HK, Delius H, Freese UK, Clarke J, Bornkamm GW. A putative transforming gene of Jijoye virus differs from that of Epstein-Barr virus prototypes. Virology. 1985;141(2):221–34. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

