Exon organization and novel alternative splicing of Ank3 in mouse heart
- PMID: 26024478
- PMCID: PMC4449188
- DOI: 10.1371/journal.pone.0128177
Exon organization and novel alternative splicing of Ank3 in mouse heart
Abstract
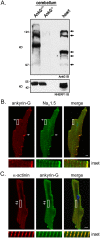
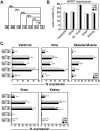
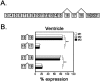
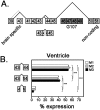
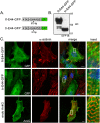
Ankyrin-G is an adaptor protein that links membrane proteins to the underlying cytoskeletal network. Alternative splicing of the Ank3 gene gives rise to multiple ankyrin-G isoforms in numerous tissues. To date, only one ankyrin-G isoform has been characterized in heart and transcriptional regulation of the Ank3 gene is completely unknown. In this study, we describe the first comprehensive analysis of Ank3 expression in heart. Using a PCR-based screen of cardiac mRNA transcripts, we identify two new exons and 28 alternative splice variants of the Ank3 gene. We measure the relative expression of each splice variant using quantitative real-time PCR and exon-exon boundary spanning primers that specifically amplify individual Ank3 variants. Six variants are rarely expressed (<1%), while the remaining variants display similar expression patterns in three hearts. Of the five first exons in the Ank3 gene, exon 1d is only expressed in heart and skeletal muscle as it was not detected in brain, kidney, cerebellum, and lung. Immunoblot analysis reveals multiple ankyrin-G isoforms in heart, and two ankyrin-G subpopulations are detected in adult cardiomyocytes by immunofluorescence. One population co-localizes with the voltage-gated sodium channel NaV1.5 at the intercalated disc, while the other population expresses at the Z-line. Two of the rare splice variants excise a portion of the ZU5 motif, which encodes the minimal spectrin-binding domain, and these variants lack β-spectrin binding. Together, these data demonstrate that Ank3 is subject to complex splicing regulation resulting in a diverse population of ankyrin-G isoforms in heart.
Conflict of interest statement
Figures



References
-
- Kordeli E, Ludosky MA, Deprette C, Frappier T, Cartaud J. AnkyrinG is associated with the postsynaptic membrane and the sarcoplasmic reticulum in the skeletal muscle fiber. Journal of cell science. 1998;111 (Pt 15):2197–207. . - PubMed
-
- Thevananther S, Kolli AH, Devarajan P. Identification of a novel ankyrin isoform (AnkG190) in kidney and lung that associates with the plasma membrane and binds alpha-Na, K-ATPase. The Journal of biological chemistry. 1998;273(37):23952–8. Epub 1998/09/03. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

